Table of Contents
What is cDNA library?
- A DNA copy that is created from messenger RNA(mRNA) through the use of reverse transcriptase enzyme is called cDNA.
- A collection of cDNA fragments that have been cloned to form a distinct vector molecule that forms part of the transcriptome of the organism . kept as a library is referred to as an cDNA library.
- The cDNA library is made up of duplicated cDNA (complementary DNA) fragments that have been inserted into a set of host cells. They form some of the transcriptomes of an organism. These fragments are stored in an “library”.
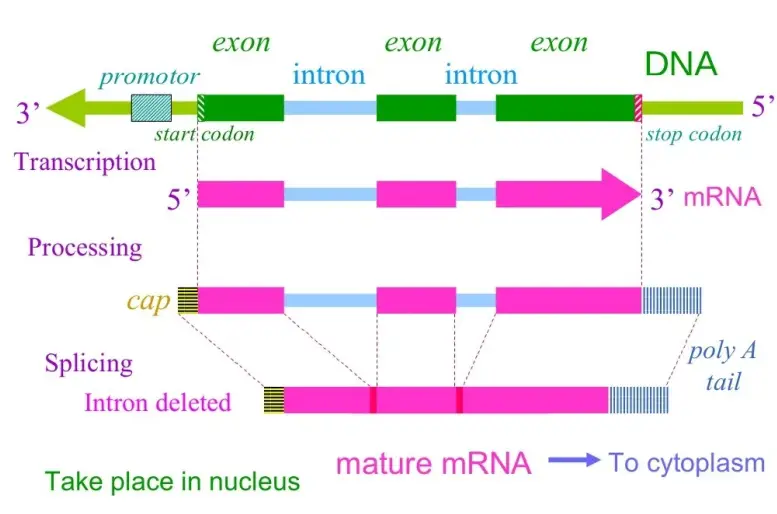
- cDNA is created by fully transcribed mRNA in the nucleus. It is only composed of the genes that are expressed in the organism. Similar tissues-specific cDNA libraries can be created.
- In eukaryotic cells, the mature mRNA has already been processed, so the cDNA that is produced does not have introns and is easily produced in a bacterial cell.
- The information found within cDNA libraries can be a powerful and effective tool, as the gene products can be identified easily however, they are not able to provide details about enhancers, introns and other regulatory elements in a genome DNA library.
Principle of cDNA library
- CDNA is produced from mRNA by the use of an enzyme called reverse transcriptase.
- In the eukaryotic cell in eukaryotic cells, there is a poly-(A) tail (consisting of an extensive sequence of Adenine Nucleotides) is the main difference between mRNA and the rRNA.
- The RNA content extracted from cells may be filtered through a chromatography column in which a strand of complementary nucleotides, such as a short uracil [poly(U) or thymidine exacts (oligo-dT) are connected to a matrix within the column.
- Its poly-(A) tail is able to attach to the oligo-DTs while the rest is removed. Evaporation using a low salt buffer lets the mRNA go through the oligo-dTs.
- After the purified mRNA has been extracted, reverse transcriptase is used to create double-stranded DNA templates from the MRNA templates.
- The newly-formed DNA-based templates can then be put into plasmids of bacteria using restriction enzymes (to cut the plasmid open) and the ligase (to stop the ends from leaking after the DNA has been annealed into the plasmid).
- Unfortunately, only a tiny percent of plasmids actually contain DNA that has been successfully integrated into the plasmid, despite a significant percentage of them being cut through restriction enzymes. A lot of plasmids remain cut or anneal to itself.
- To identify plasmids which have the DNA that has been successfully inserted, an additional antibiotic resistance gene is typically inserted alongside the DNA. The bacteria is then raised in a media containing this antibiotic.
- All bacteria that carry plasmids which have been able to successfully absorb the DNA (and thus the antibiotic resistance) can then live and all other bacteria will end up disappearing.
- The colonies that survive are created in order to replicate the plasmid using the DNA. It is later isolated and removed and purified. This is why these kinds of libraries are referred to as cDNA also known as complementary DNA since it’s a complementary DNA strand that is linked to the mRNA being studied.
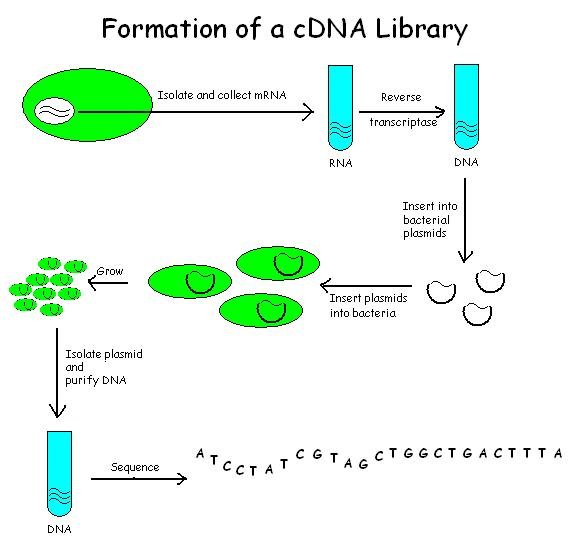
Process involved in the construction of cDNA library
1. Extraction of mRNA from the eukaryotic cell
- The mRNA that is derived from the remaining RNAs is gathered and then purified.
- There are many other options to purify RNA such as the trizol extract and purification of columns.
- By utilizing oligomeric dT nucleotide-coated resins, column purification is conducted where only the poly-A tail’s mRNA can bind.
- All the remaining RNAs have been eluted.
- The mRNA is then eluted with an eluting buffer as well as some heat to break up the mRNA strands from oligo-dT.
2. cDNA construction
In the process of creating cDNAs, there’s different approaches. They are described as follows:
a. The RNAse method
- Principle: Through using reverse transcriptase, a DNA complement is synthesized, forming an RNA-DNA duplex. The DNA strand of RNA is cut before being replaced by DNA.
Step I: Annealing:
- A chemically synthesized oligo-dT primer is annealed onto the 3′ polyA-tail of RNA. The primer can be found to be 10 to 15 residues long.
- In the presence of reverse transcriptase and deoxyribonucleotides, it primes the synthesis of the first DNA strand. It results in a RNA:DNA duplex.
Step II: Replacing RNA strand with DNA strand:
- The RNA string is replaced with DNA strand with the aid of the enzyme RNAse H.
- The enzyme RNase removes the RNA from the RNA:DNA duplex. The DNA strand left behind acts as a template, as does another DNA strand that is synthesized by the DNA Polymerase II.
b. The self-priming method
- In this way the oligo-dT primer is attached to the polyadenylate tail of mRNA in order to initiate the first DNA strand synthesise against to the mRNA.
- This cDNA as it is formed tends to fold over it, temporarily, and form hairpin loops.
- This causes the second self-priming of the strand.
- This loop must be cleaved with a single-strand-specific nuclease, e.g., SI nuclease, after the synthesis of the second DNA strand to allow insertion into the cloning vector.
- There is a major disadvantage to this method. At the 5′-end of the clone, cleavage using SI nuclease causes the loss of a specific number of sequences.
c. Land et al. strategy
- The cDNA is linked to the Cytidine residues through the enzyme terminal transferase in the first-strand synthesis process, and is primed using an oligo-dT primer , as is the case.
- To create a synthetic oligo-dG based primer, this synthetic oligo-dC tail is used to serve as an annealing spot that allows the 2nd strand of DNA to synthesize.
d. Homopolymer tailing
- The terminal transferase enzyme which can polymerize nucleotides and convert them into the 3′-hydroxyl of DNA as well as RNA is employed in this process.
- To make an RNA DNA hybrid the synthesis process of the first DNA strand takes place exactly as before.
- To add nucleotide tails onto the 3″ ends of DNA and RNA Strands, terminal transferase is used and one deoxyribonucleotide is employed.
- The consequence is that at its 3″ end the DNA strand has a well-known sequence. DCTP as well as dATP is typically employed.
- A complementing an oligomer (chemically synthesized) is now made annealed and utilized as a primer for the process of synthesis of the second Strand.
- In order to assist in the cloning of the resulting double-stranded DNA the oligomer (and it is also the one that is used for the first strand synthesis) can also incorporate the restriction site.
e. Rapid amplification of cDNA ends
The RACE techniques are divided into 5’RACE and 3’RACE in accordance with the end of the cDNA that we are interested in.
(i). 3’ RACE
Reverse transcriptase synthesis of the initial DNA strand occurs with a modified oligo-dT primer used in this type of RACE. This primer is the extension of a specific adaptor sequence that is followed by an extended oligo DT. The initial strand synthesis is followed by another strand synthesis which utilizes a primer which is internal to the coding sequence of the interest. This is followed by PCR, which employs
- The same internal primer.
- The sequence of adaptors (i.e. not including the adaptor’s oligo-dT). While it is feasible to use a basic oligo-dT-based primer in theory instead of adaptor-oligo-dT-adaptor combination however, the low melting temperature may interfere with the subsequent PCR rounds that are required for an oligodT primer.
(ii). 5’ RACE
The initial cDNA is a strand in RACE synthesized by the enzyme reverse transcriptase as well as an unincorporated primer from the codon sequence. It removes the non-integrated primer and then tails the cDNA in strands that contain the oligo-dA. By using an adaptor-oligodT primer the second cDNA strand can later be synthesized. The double-stranded molecule that results from this process are later subjected to PCR
- A primer nestled within the coding region
- When the final PCR is performed an nested primer is employed to increase the specificity. Because of the low melting point of a standard oligo-dT primer the adaptor sequence is employed during the PCR process, just like in the 3’RACE above. There are a variety of kits accessible commercially to use RACE.
3. cDNA cloning
a. Linkers
- At the final stage, techniques used by RNaseH or homopolymer tailing produce the double-stranded, blunt-ended cDNA molecules.
- The vector molecules have to be attached to them.
- This is possible through the use of blunt-ended ligation or digestion with the enzyme rela-evant and the litigation process into the vector as well as by adding linkers.
b. Incorporation of restriction sites
- The homopolymer tailing process can be modified by using primers that can be modified to accommodate the restrictions.
- The 3 ‘ end of the initial cDNA string, which was synthesized recently it is joined by C’s.
- An oligo-dG synthesis primer, preceded by a sail-site in the oligonucleotide’s shorter double-stranded area, is utilized for second-strand synthesizing.
- The application of an oligonucleotide with two-stranded regions is required to accomplish this.
- The oligonucleotides that are produced are made by synthesizing two strands before they are allowed to anneal one another.
c. Homopolymer Tailing of cDNA
- Another suggestion is to reuse terminal transferase.
- Treatment using terminal transferase as well as dCTP of double-stranded cDNA with blunt ends can result in the polymerization of many C residues (typically 20 or more) in 3′ hydroxyls at each end.
- The transferase at the end as well as the dGTP processing of the vector results in the addition of several G residues on the edges of the vector. It is also possible to utilize the dATP or dTTP method in different ways.
- There is now a way to join both the vector and cDNA the base-paired region can be large enough that the need for DNA ligase is not required.
- There could be gaps , not nicks around areas around the vector insert but after the recombinant molecules are introduced into the host, they are repaired through the physiological processes.
How to screen cDNA library?
- Immobilize members of the library on a membrane of nylon and alter them until they’re single-stranded
- Make a radiolabelled probe and then denature it so that it single-stranded
- Hybridize the probe with the library of the clones
- Clean the probe of any excess and expose the film of X-rays
- Determine the positive clone’s identity and then analyze
The process of hybridization is carried out at an non-stressful temperature which guarantees that the probe can bind to any clone with the same sequence. In addition, non-specific hybridization may occur as some clones have a limited, but not significantly similarity in comparison to that probe. The washing procedure is conducted at an high temperature that is enough to clean the probe of all clones it has been bound in a non-specific way. It is crucial to ensure that the temperature isn’t enough to wash the probe of clones with sequences that are comparable or similar to the probe. Thus, consideration of the origin for the probe (homologous or heterologous) determines how high washing process is carried out.
Advantages of cDNA library
There are two main benefits to the cDNA library.
- In the beginning, it is enhanced by fragments of genes that are being transcribed.
- Introns are not the only way to alter the cloned sequences should the objective be to make a eukaryotic protein in bacteria, introns could be a challenge, because the majority of bacteria lack a method to eliminate the introns.
Disadvantages of cDNA library
- A cDNA library is a disadvantage in that it contains only sequences that are found inside mature mRNA.
- Introns are not present, nor are other sequences that are altered during transcription. Sequences that aren’t transcribed into RNA, like enhancers and promoters are not included in a collection of cDNA.
- It is important to note that only certain gene sequences that are expressed inside the organ from which DNA has been isolated make up the cDNA library.
- Additionally to that, in an cDNA collection, the amount of a particular DNA sequence is determined by the amount of the relevant MRNA within the tissue of interest.
- Contrary to this when you look at a genome-wide DNA library, virtually all genes are found at an equal frequency.
Application of cDNA library
- CDNA libraries are typically employed when reproducing genomes of eukaryotic origin, because the amount of data is decreased to eliminate the huge number of non-coding regions in the library.
- CDNA libraries can be used to express the eukaryotic genes in prokaryotes. Prokaryotes don’t have introns within their DNA, so they do not have any enzymes that could cut it out during transcription. CDNA are intron-free and thus can be expressed by prokaryotic cells.
- CDNA libraries are the most valuable in reverse genetics , where the additional genomic information is of less use.
- Additionally, it can be helpful in the future to isolate the gene that encodes for that particular mRNA.
- Discovering novel genes.
- Cloning of full-length CDNA molecules for the in vitro research into the function of genes. Research into the diversity of the mRNAs are expressed in various tissues or cells.
- The study of alternative splicing within different tissues or cells.
FAQ
what is cdna library?
A cdna library is the collection of genetic data for organisms, usually for human beings. The genome project started in 1990 and completed in 2003. The Human Genome Project sequenced the entire DNA sequence of the human species. Scientists discovered about 30 million genes.
cdna library uses which enzyme?
CdnA library uses DNA polymerase I (Klenow fragment). The Klenow fragment has high specificity for dNTPs and is able to incorporate them into the growing strand of DNA. It also exhibits strong exonuclease activity. This makes it useful for sequencing applications.
What is the starting material for making a cdna library?
A cDNA library is made using mRNA instead of DNA as the starting material. The mRNA can be extracted from cells of specific tissues from the organism of interest. The “c” in cDNA stands for copy because a double stranded DNA copy is made from a mRNA .
References
- Zhang, P., & Min, X. J. (2005). EST Data Mining and Applications in Fungal Genomics. Applied Mycology and Biotechnology, 33–70. doi:10.1016/s1874-5334(05)80004-8
- Xu, Y., Zhou, J., Liu, Q. et al. Construction and characterization of a high-quality cDNA library of Cymbidium faberi suitable for yeast one- and two-hybrid assays. BMC Biotechnol 20, 4 (2020). https://doi.org/10.1186/s12896-020-0599-2
- https://www.slideshare.net/ramsdude/complementary-dna-cdna-libraries-28566053
- https://www.slideshare.net/NushratJahan4/cdna-library-preparation-ppt-for-jamil-sir
- https://link.springer.com/book/10.1385/1592593593
- https://link.springer.com/book/10.1385/089603383X
- https://experiments.springernature.com/articles/10.1385/1-59259-283-X:349
- https://www.takarabio.com/products/mrna-and-cdna-synthesis/cdna-synthesis-kits/library-construction-kits
- https://www.bionity.com/en/encyclopedia/CDNA_library.html
- https://www.britannica.com/science/cDNA-library
- https://www.creative-biogene.com/support/cdna-library-construction-protocol.html
- https://www.sciencedirect.com/science/article/pii/B9780128205952000023?via%3Dihub
- https://cdn.technologynetworks.com/ep/pdfs/cdna-library-construction-1.pdf
- https://bmcbiotechnol.biomedcentral.com/articles/10.1186/s12896-020-0599-2
- https://www.onlinebiologynotes.com/cdna-library-process-of-construction-of-cdna-library-advantages-and-disadvantages/

A cDNA library is made using mRNA instead of DNA as the starting material. The mRNA can be extracted from cells of specific tissues from the organism of interest. The “c” in cDNA stands for copy because a double stranded DNA copy is made from a mRNA .