Table of Contents
Coronavirus
- Coronaviruses are a family of viruses that can cause respiratory illnesses in humans and animals.
- The virus was first identified in 1937 and has since been responsible for several outbreaks, including the severe acute respiratory syndrome (SARS) outbreak in 2003 and the Middle East respiratory syndrome (MERS) outbreak in 2012.
- The current strain of coronavirus causing the COVID-19 pandemic is known as SARS-CoV-2.
- It is primarily transmitted through respiratory droplets when an infected person talks, coughs, or sneezes.
- Symptoms can include fever, cough, fatigue, and difficulty breathing.
- The virus can be severe in some cases, particularly in older adults and those with underlying health conditions.
- There are currently several vaccines available to prevent COVID-19, and treatments are available to manage symptoms and reduce the risk of severe illness.
- Public health measures such as wearing masks, social distancing, and frequent hand washing can also help to prevent the spread of the virus.
- The pandemic has had a significant impact on global health, economies, and societies.
- Efforts are ongoing to control the spread of the virus through vaccination campaigns, testing, and contact tracing.
- New variants of the virus continue to emerge, leading to concerns about their potential impact on the efficacy of vaccines and the effectiveness of public health measures.
- COVID-19 has highlighted the importance of preparedness for future pandemics and the need for investment in public health infrastructure and research.
- The World Health Organization (WHO) has declared the COVID-19 pandemic a Public Health Emergency of International Concern (PHEIC).
- COVID-19 has led to unprecedented levels of collaboration among researchers, healthcare professionals, and governments worldwide.
- The pandemic has also had significant social and psychological impacts, including increased rates of anxiety, depression, and social isolation.
Characteristics of COVID-19
- COVID-19 is characterized by fever and a raspy cough.
- Most patients exhibit bilateral pneumonia.
- SARS-CoV-2 is more likely to affect elderly men with multiple comorbidities.
- Blood samples of COVID-19 patients reveal leucopenia and lymphopenia.
- ICU patients’ plasma levels of cytokines including IL2, IL7, IL10, GSCF, IP10, MCP1, MIPA, and TNFa are elevated.
- There are three levels of severity for COVID-19: moderate, severe, and critical.
- There have been reports of asymptomatic instances, but the majority of patients eventually develop the disease.
- COVID-19 is associated with myocardial damage, neurological complications, stroke, digestive symptoms, liver damage, hypercoagulability, and thrombotic complications.
- Patients in a critical condition may develop ARDS, metabolic acidosis, septic shock, coagulopathy dysfunction, and multiple organ functional failure.
- About 26.1% of patients are admitted to the ICU because of COVID-19-related complications.
- With correct diagnosis and treatment, the majority of patients have a favorable prognosis.
- The prognosis for elderly patients and those with underlying diseases is poorer.
Structure of Coronavirus
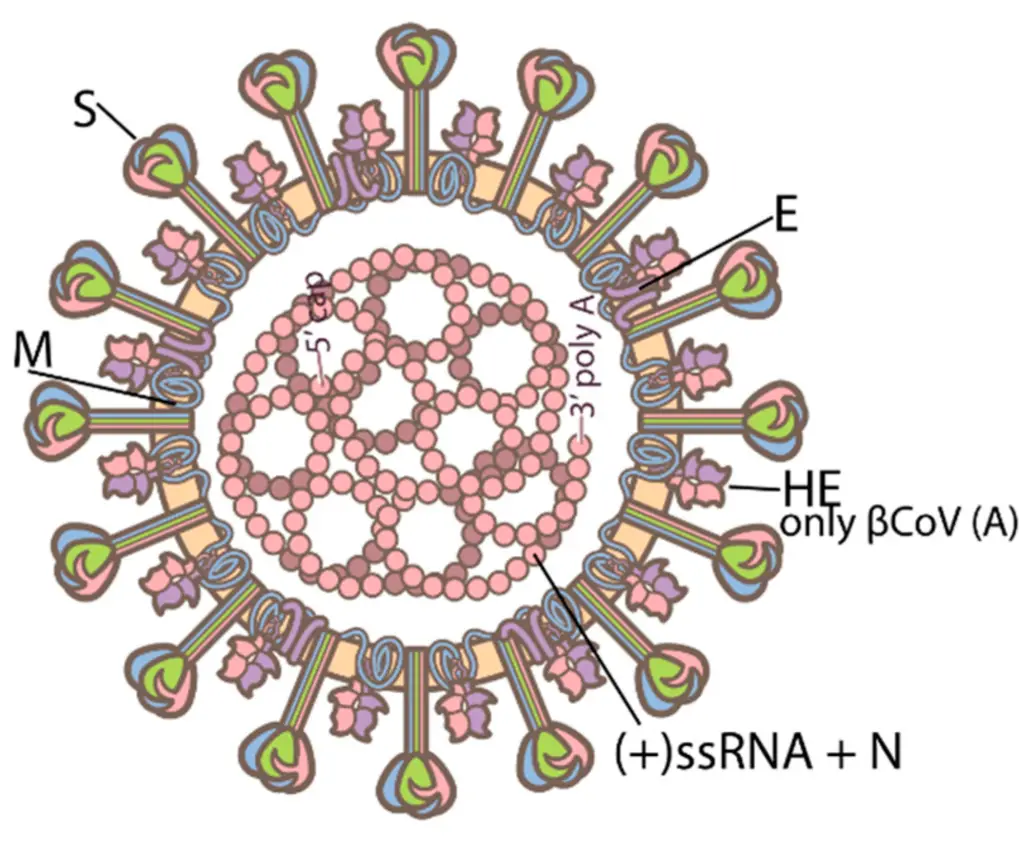
- Coronaviruses belong to the subfamily Coronavirinae within the family Coronaviridae, which consists of four genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus. The genome of CoVs (27–32 kb) is a single-stranded positive-sense RNA (+ssRNA) that is greater than that of any other RNA virus.
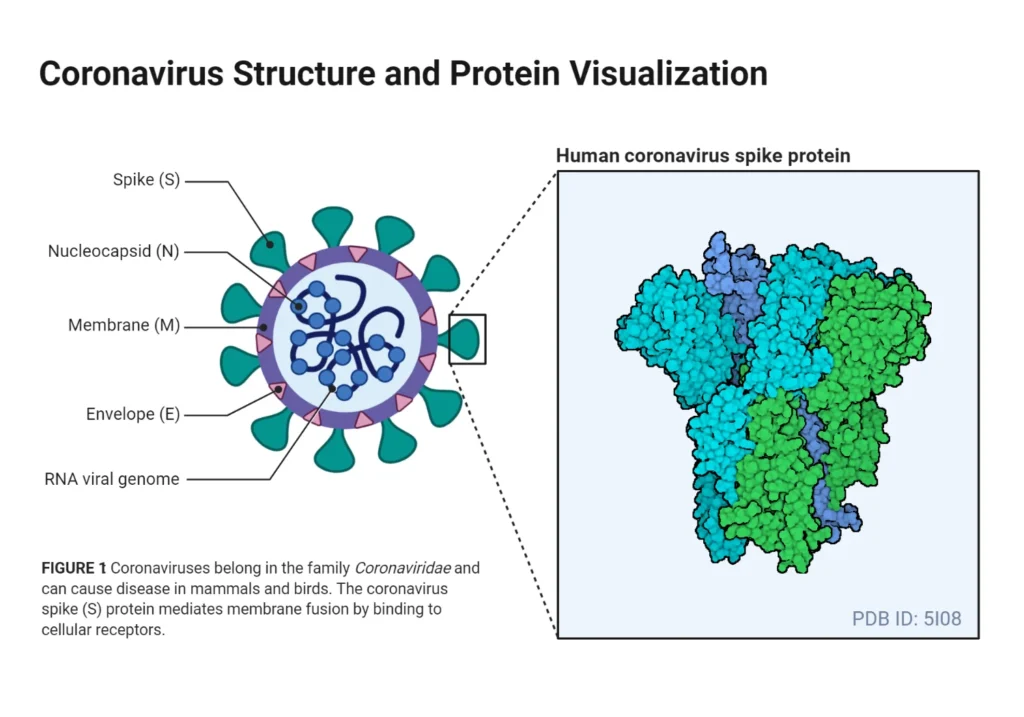
- The nucleocapsid protein (N) formed the capsid outside of the genome, and the genome is further encased by an envelope composed of three structural proteins: membrane protein (M), spike protein (S), and envelope protein (E).
- As a coronavirus, the recently sequenced genome of SARS-CoV-2 is approximately 29.9 kilobytes in size. The SARS-CoV-2 virus is composed of four structural proteins (S, E, M, and N) and sixteen nonstructural proteins (nsp116).
- RNA processing and replication are mediated by Nsp1. The survival signaling pathway of the host cell is modulated by Nsp2. It is believed that Nsp3 disassembles the translated protein. Nsp4 contains transmembrane domain 2 (TM2) and modifies ER membranes.
- Nsp5 is involved in the replication polyprotein synthesis process. Nsp6 is a presumptive transmembrane domain. The combination of nsp12 and template-primer RNA was substantially enhanced in the presence of nsp7 and nsp8. ssRNA-binding protein is Nsp9’s function. Nsp10 is essential for the methylation of the caps of viral mRNAs.
- Nsp12 contains RNA-dependent RNA polymerase (RdRp), a crucial component of coronavirus replication/transscription. Nsp13 bonds to ATP, and the zinc-binding domain in nsp13 is involved in replication and transcription. Nsp14 is a domain for exoribonuclease proofreading. Endoribonuclease activity of Nsp15 is dependent on Mn(2+).
- Naqvi et al. (2020) identified Nsp16 as a 2′-Oribose methyltransferase. One study demonstrates that NSP-mediated effects on splicing, translation, and protein trafficking serve to suppress host defenses. NSP16 inhibits mRNA splicing upon SARSCoV-2 infection by binding the mRNA recognition domains of the U1 and U2 snRNAs.
- NSP1 inhibits mRNA translation by binding to 18S ribosomal RNA in the ribosome’s mRNA entry channel. NSP8 and NSP9 bind to the 7SL RNA, which localizes at the Signal Recognition Particle, to interfere with protein trafficking to the cell membrane. On the basis of their structures, the following SARSCoV-2 proteins may serve as potential antiviral drug targets.
Genome of Coronavirus
- Coronaviruses have a single-stranded, positive-sense RNA genome. Coronavirus genomes range in size from 26,4 to 31,7 kilobases.
- It has one of the largest genomes among RNA viruses. The genome has a 3′ polyadenylated tail and a 5′ methylated end.
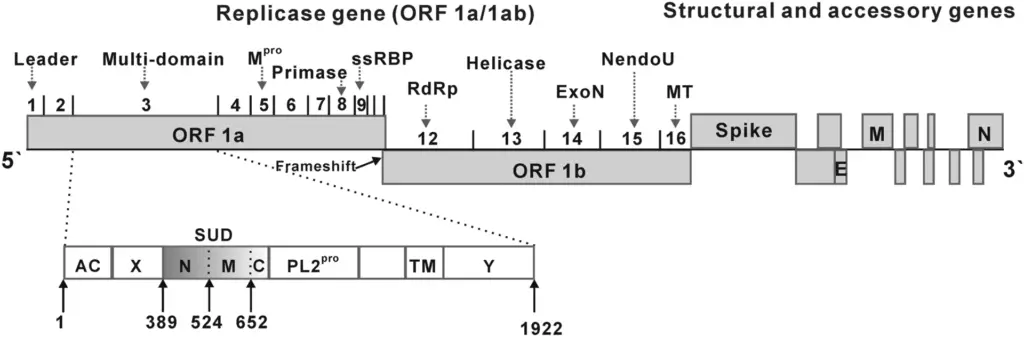
- The organization of the coronavirus genome is 5′-leader-UTR replication case (ORF1ab).-spike (S)-envelope (E)-membrane (M)-nucleocapsid (N)3’UTR poly (A) tail.
- The replicase polyprotein (pp1ab) is encoded by the open reading frames 1a and 1b, which span the first two-thirds of the genome. Self-cleavage of the replicase polyprotein yields 16 nonstructural proteins (nsp1–nsp16).
- The four main structural proteins, spike, envelope, membrane, and nucleocapsid, are encoded in the later reading frames.
- These reading frames are interspersed with the reading frames for the ancillary proteins. The number and function of accessory proteins vary depending on the specific coronavirus.
Epidemiology of Coronavirus
The novel coronavirus SARS-CoV-2 is the cause of the 2019 coronavirus disease (COVID-19). The virus was identified for the first time in December 2019 in Wuhan, China, and has since spread worldwide, resulting in a pandemic. Here are some essential aspects of the epidemiology of COVID-19:
- The virus is transmitted predominantly through respiratory droplets when an infected person coughs or sneezes, or by touching an infected surface and then touching one’s face.
- The estimated incubation period for the virus is between 2 and 14 days, during which asymptomatic infected individuals can transmit the virus to others.
- The virus has a higher transmission rate than seasonal influenza, but the case fatality rate varies significantly based on age, sex, and underlying health.
- Those with underlying health conditions, such as heart disease, diabetes, or a compromised immune system, are at a greater risk for severe illness and mortality.
- There are disparities in how the virus has affected various communities, with certain racial and ethnic groups suffering from higher infection, hospitalization, and mortality rates.
- The pandemic has had a significant impact on global health, economies, and daily life, with many countries instituting lockdowns, travel restrictions, and mask mandates to halt the virus’s spread.
- Vaccines have been developed and approved for emergency use, with ongoing efforts to increase their global distribution and accessibility.
- Virus variants have emerged and continue to evolve, with some variants being more contagious or capable of causing more severe illness.
- Important for controlling the spread of the virus and preventing future epidemics are continued surveillance, testing, and contact tracing.
Replication of Coronavirus
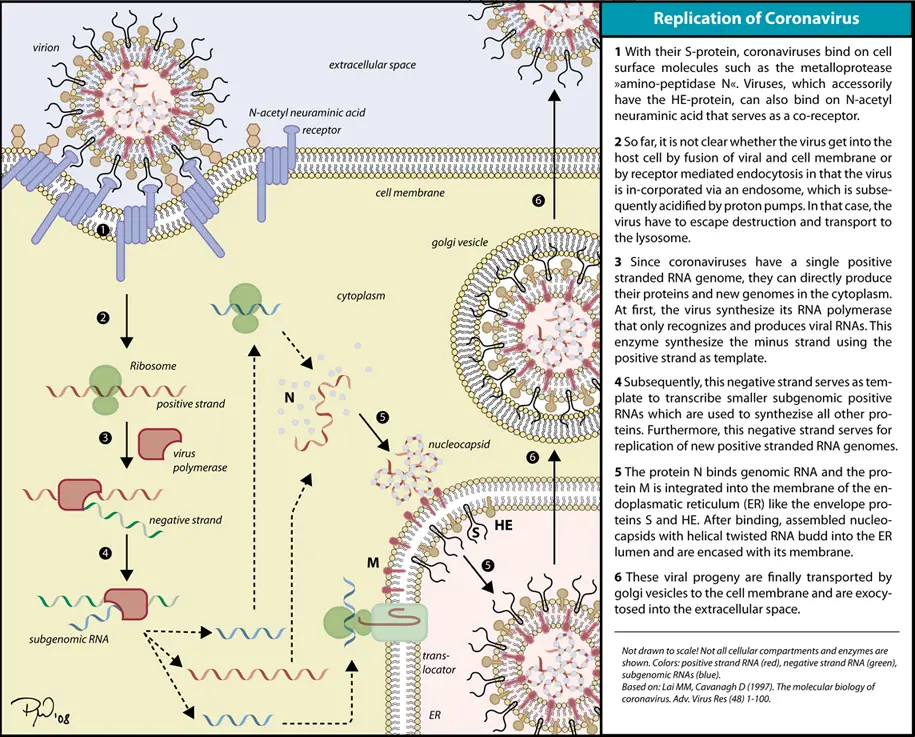
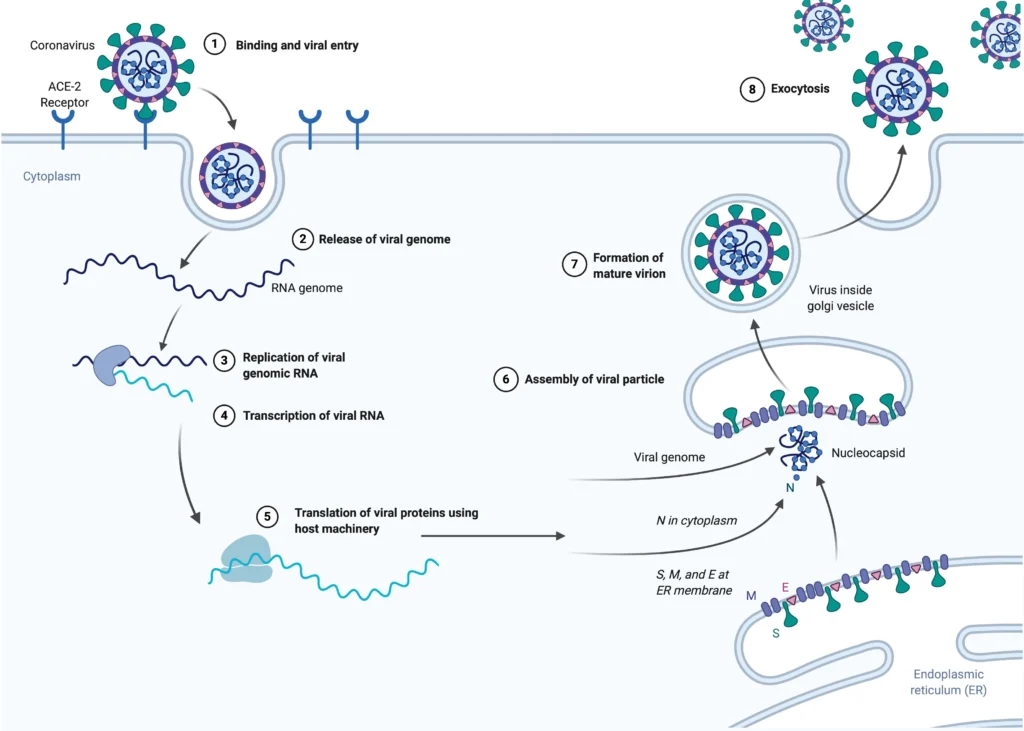
1. Cell entry
- When the viral spike protein binds to its complementary host cell receptor, infection begins. A protease of the host cell cleaves and activates the receptor-attached spike protein after attachment. Depending on the protease present in the host cell, cleavage and activation permit the virus to enter the host cell via endocytosis or direct fusion of the viral envelope with the host membrane.
- Coronaviruses can penetrate cells either by fusing their lipid envelope to the cell membrane on the cell surface or by endocytosis.
2. Genome translation
- The virus particle is uncoated upon entry into the host cell, and its genome enters the cytoplasm. The RNA genome of the coronavirus has a 5′ methylated cap and a 3′ polyadenylated tail, allowing it to operate as a messenger RNA and be directly translated by the ribosomes of the host cell.
- The initial overlapping open reading frames ORF1a and ORFb of the virus genome are translated by the host ribosomes into two large overlapping polyproteins, pp1a and pp1b.
- The larger polyprotein pp1ab results from a -1 ribosomal frameshift caused by a slippery sequence (UUUAAAC) and an RNA pseudoknot downstream at the end of open reading frame ORF1a. The ribosomal frameshift permits the uninterrupted translation of ORF1a and ORFb.
- Polyproteins have their own proteases, PLpro (nsp3) and 3CLpro (nsp5), which cleave them at distinct locations. 16 nonstructural proteins (nsp1 to nsp16) are generated by the cleavage of polyprotein pp1ab. Product proteins consist of RNA-dependent RNA polymerase (nsp12), RNA helicase (nsp13), and exoribonuclease (nsp14), among others.
3. Replicase-transcriptase
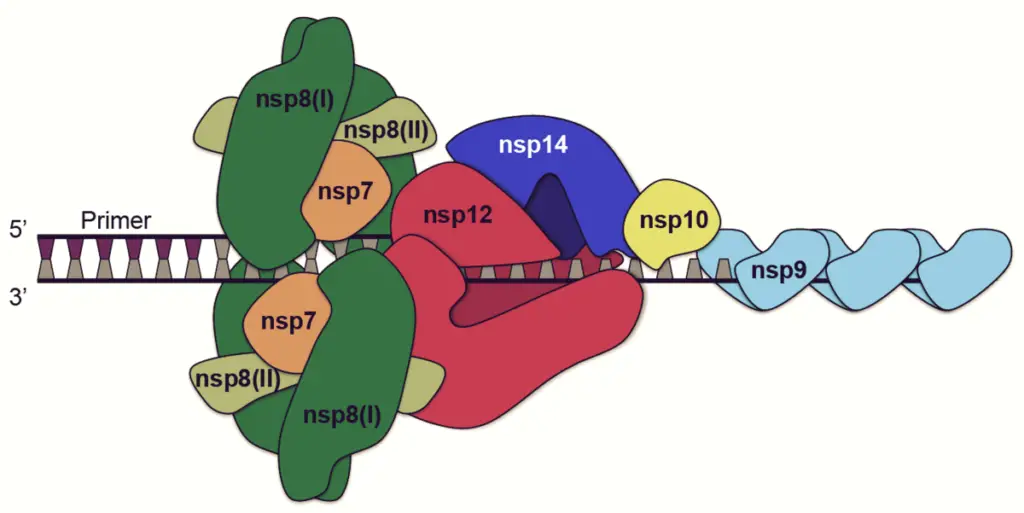
- A number of the nonstructural proteins coalesce to produce a multi-protein replicase-transcriptase complex (RTC). RNA-dependent RNA polymerase (RdRp) is the principal replicase-transcriptase protein. It participates directly in the replication and transcription of RNA from an RNA strand.
- The complex’s other nonstructural proteins aid in the replication and transcription processes. The exoribonuclease nonstructural protein, for example, increases replication fidelity by performing a proofreading function that RNA-dependent RNA polymerase lacks.
- Replication – One of the complex’s primary functions is to replicate the viral genome. RdRp mediates directly the formation of negative-sense genomic RNA from positive-sense genomic RNA. Following this, positive-sense genomic RNA is replicated from negative-sense genomic RNA.
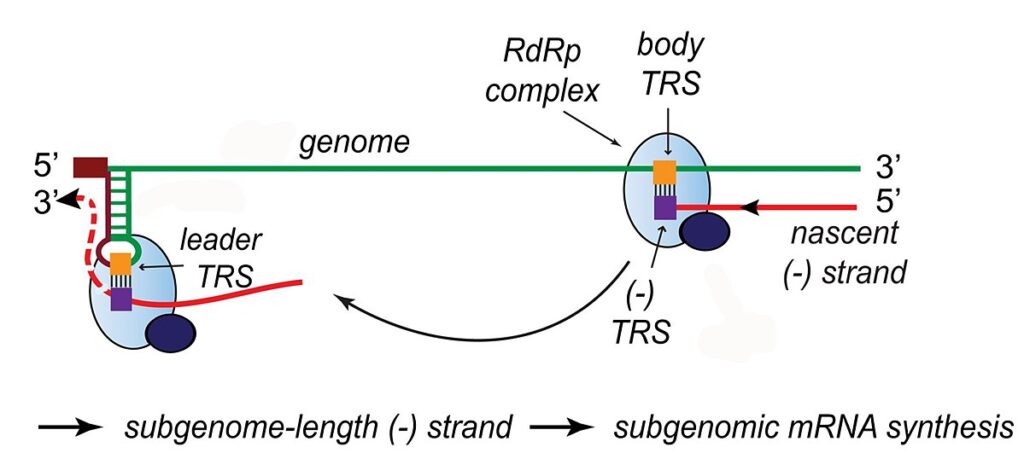
- Transcription – The complex’s other essential function is to transcribe the viral genome. RdRp directly mediates the synthesis of subgenomic RNA molecules with negative sense from genomic RNA with positive sense.
- The subsequent step is the transcription of these negative-sense subgenomic RNA molecules into their positive-sense mRNA counterparts. The subgenomic mRNAs form a “nested set” with a common 5′-head and a 3′-end that is partially duplicated.
- When at least two viral genomes are present in the same infected cell, the replicase-transcriptase complex is also capable of genetic recombination.
- RNA recombination appears to be a significant driving force in determining genetic variability within a coronavirus species, the capability of a coronavirus species to jump from one host to another and, infrequently, in determining the emergence of novel coronaviruses.
- Uncertainty surrounds the precise mechanism of recombination in coronaviruses, but it appears to entail template switching during genome replication.
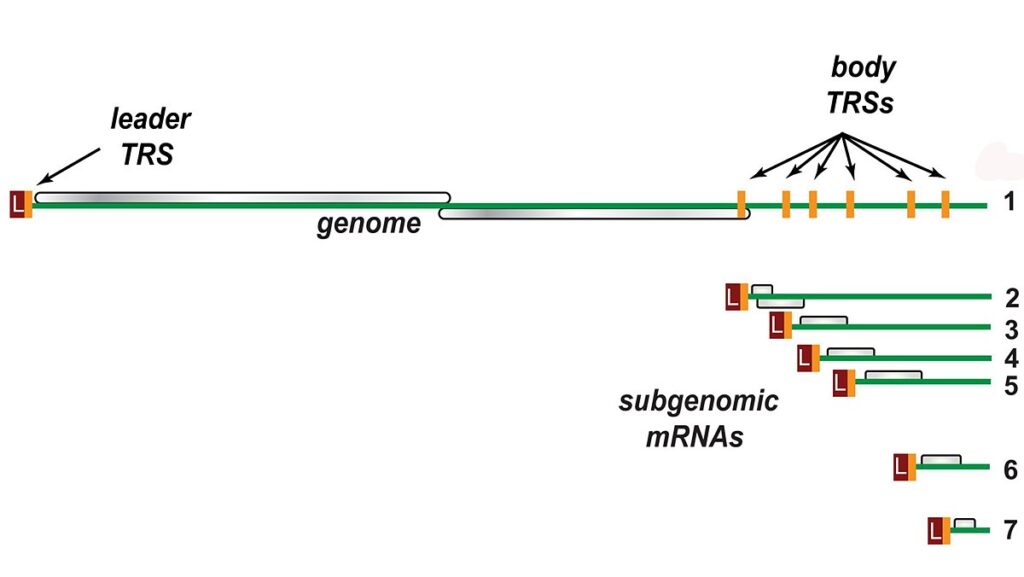
4. Assembly and release
- The replicated positive-sense genomic RNA serves as the virus’s genome. After the initial overlapping reading frame, the mRNAs are gene transcripts of the final third of the virus genome.
- The ribosomes of the host translate these mRNAs into structural proteins and numerous accessory proteins.
- Translation of RNA takes place within the endoplasmic reticulum. The structural proteins S, E, and M of the virus travel along the secretory pathway to the Golgi intermediate compartment.
- Following their binding to the nucleocapsid, the M proteins direct the majority of protein-protein interactions required for the assembly of viruses. Viral offspring are then expelled from the host cell via exocytosis and secretory vesicles. Once viruses are released, they can infect other host cells.
Pathogenesis of Coronavirus
- SARS-CoV-2 is structurally and phylogenetically similar to SARS-CoV and MERS-CoV, and consists of four major structural proteins: spike (S), envelope (E) glycoprotein, nucleocapsid (N), and membrane (M) protein, as well as 16 nonstructural proteins and 5 to 8 accessory proteins.
- The surface spike (S) glycoprotein, which resembles a crown, is located on the outer surface of the virion and undergoes cleavage into an amino (N)-terminal S1 subunit, which facilitates virus incorporation into the host cell, and a carboxy (C)-terminal S2 subunit containing a fusion peptide, a transmembrane domain, and cytoplasmic domain, which is responsible for virus-cell membrane fusion.
- The S1 subunit is further subdivided into a receptor-binding domain (RBD) and an N-terminal domain (NTD), which facilitates viral entry into the host cell and is a potential target for neutralization by antisera or vaccines.
- As a binding site for the human angiotensin-converting enzyme 2 (ACE2) receptors, the RBD is a fundamental peptide domain in the pathogenesis of infection. As previously hypothesized, inhibition of the renin-angiotensin-aldosterone system (RAAS) does not increase the risk of hospitalization for COVID-19 and severe disease.
- By binding the SARS-CoV-2 spike or S protein (S1) to the abundant ACE2 receptors on respiratory epithelium such as type II alveolar epithelial cells, SARS-CoV-2 gains access to host cells. ACE2 receptors are also expressed by the upper esophagus, enterocytes from the ileum, myocardial cells, proximal tubular cells of the kidney, and urothelial cells of the bladder, in addition to the respiratory epithelium.
- The process of viral attachment is followed by the priming of the spike protein S2 subunit by the host transmembrane serine protease 2 (TMPRSS2), which facilitates cell entry, viral replication endocytosis, and the assembly of virions.
- In conclusion, the spike RBD enables binding to the ACE2 receptor in lungs and other tissues. The spike protein of an amino acid site (polybasic site) allows furin (protease) to metabolize the amino acid site functionally. This process allows for the exposure of the fusion sequences and, consequently, the fusion of the viral and cell membranes, which is required for the virus to enter the cell.
Effect of SARS-CoV-2 on the Respiratory System/ Pathogenesis of SARS-CoV-2-induced Pneumonia
- COVID-19 primarily targets the respiratory and vascular systems caused by SARS-CoV-2.
- The pathogenesis of SARS-CoV-2 induced pneumonia has two phases: early and late.
- The early phase is caused by viral replication resulting in tissue damage.
- The late phase is characterized by the immune system’s response with the recruitment of T lymphocytes, monocytes, and neutrophils, which release cytokines such as TNF α, GM-CSF, IL-1, IL-6, IL-1β, IL-8, IL-12 and IFN-γ.
- Severe COVID-19 leads to an overactivation of the immune system, causing a cytokine storm that releases high levels of cytokines, especially IL-6 and TNF-α, causing a local and systemic inflammatory response.
- Increased vascular permeability and subsequent development of pulmonary edema in severe COVID-19 patients are explained by multiple mechanisms, including endotheliitis, dysregulation of the RAAS, activation of the kallikrein-bradykinin pathway, and enhanced epithelial cell contraction.
- Binding of SARS-CoV-2 to the TLR induces the release of pro-IL-1β, which is cleaved into the active mature IL-1β that mediates lung inflammation until fibrosis.
Effect of SARS-CoV-2 on Extrapulmonary Organ Systems
SARS-CoV-2 primarily affects the respiratory system, but it also has the potential to cause problems in the gastrointestinal (GI), hepatobiliary, cardiovascular (CV), renal (R), and central nervous systems. Direct viral toxicity, ischemic injury due to vasculitis, thrombosis, or thrombo-inflammation, immune dysregulation, and renin-angiotensin-aldosterone system (RAAS) dysregulation are all possible explanations for SARS-CoV-2-induced organ dysfunction.
- Cardiovascular system (CVS): Although the precise mechanism of COVID-19’s cardiac involvement is unknown, it is probable that it is multifactorial. Myocardial cells also express ACE2 receptors, suggesting that SARS-CoV-2 exerts direct cytotoxicity on the myocardium, leading to myocarditis. IL-6 and other proinflammatory cytokines can cause vascular inflammation, myocarditis, and cardiac arrhythmias. Acute coronary syndrome (ACS) is a well-recognized cardiac manifestation of COVID-19 and is likely due to multiple factors, including but not limited to COVID-19-associated hypercoagulability, the release of proinflammatory cytokines, worsening of preexisting severe coronary artery disease, stress cardiomyopathy, and associated hemodynamic derangement which may reduce coronary blood flow, reduced oxygen supply resulting in the destabilization of coronary plaque microscopic structures, and associated.
- Hematological: The SARS-CoV-2 virus has a substantial impact on the hematological and hemostatic systems. Unknown is the mechanism underlying leukopenia, one of the most common laboratory abnormalities associated with COVID-19. Several hypotheses have been proposed, including ACE 2-mediated annihilation of lymphocytes by direct virus invasion, lymphocyte apoptosis due to proinflammatory cytokines, and possible virus invasion of lymphatic organs. Thrombocytopenia is uncommon in COVID-19 and is likely caused by a combination of factors, including virus-mediated suppression of platelets, formation of autoantibodies, and activation of the coagulation cascade, resulting in platelet consumption. Thrombocytopenia and neutrophilia are regarded as defining characteristics of severe illness. Although it is well known that COVID-19 is associated with a state of hypercoagulability, the exact mechanisms that contribute to the activation of the coagulation system remain unknown and are likely a result of the cytokine-induced inflammatory response. The pathogenesis of this hypercoagulability is multifactorial and is likely caused by direct viral-mediated damage or cytokine-induced injury of the vascular endothelium, resulting in the activation of platelets, monocytes, and macrophages, and the increased expression of tissue factor, von Willebrand factor, and Factor VIII, which leads to the production of thrombin and fibrin clot formation. Other proposed mechanisms include putative prothrombotic sequelae induced by mononuclear phagocytes, derangements in the renin-angiotensin system (RAS) pathways, and complement-mediated microangiopathy.
- Central Nervous System (CNS): Emerging evidence of ACE2 receptors in human and mouse brains implicates the possibility of SARS-CoV-2 infection of the brain.[62] SARS-CoV-2 may infiltrate the central nervous system via transsynaptic transmission across infected neurons via the olfactory nerve, infection of vascular endothelial cells, or migration of leukocytes across the blood-brain barrier.
- Gastrointestinal (GI) Tract: The pathogenesis of GI manifestations of COVID-19 is unknown and likely multifactorial due to a number of potential mechanisms, such as direct ACE 2-mediated viral cytotoxicity of the intestinal mucosa, cytokine-induced inflammation, gut dysbiosis, and vascular abnormalities.
- Hepatobiliary: Although the pathogenesis of liver injury in COVID-19 patients is unknown, hepatic injury in COVID-19 is likely multifactorial and is explained by a number of mechanisms alone or in combination, such as ACE-2-mediated viral replication in the liver, direct virus-mediated damage, hypoxic or ischemic injury, immune-mediated inflammatory response, drug-induced liver injury (DILI), or worsening of preexisting liver disease.
- Renal: The pathogenesis of COVID-19-associated kidney injury is unknown and is likely multifactorial, involving a single or a combination of multiple factors, such as direct cytotoxic injury from the virus, imbalance in the RAAS, cytokine-induced hyperinflammatory state, microvascular injury, and the prothrombotic state associated with COVID-19. Other factors, including associated hypovolemia, potential nephrotoxic agents, and nosocomial sepsis, may also contribute to kidney damage. During the early phase of the pandemic, a seven-month study conducted by Ziemba et al. revealed that the mortality rate per 1,000 ESRD patients during the pandemic exceeded the expected death rate per 1,000 ESRD patients based on data from previous years preceding the outbreak.
Transmission of Coronavirus
- The primary mode of transmission of SARS-CoV-2 is exposure to infectious respiratory secretions from close contact or droplet transmission from presymptomatic, asymptomatic, or symptomatic hosts.
- In addition to aerosol-generating procedures, airborne transmission has also been linked to the spread of COVID-19. Emerging and being evaluated are data implicating airborne transmission of SARS-CoV-2 in the absence of aerosol-generating procedures. Nonetheless, this mode of transmission is not universally accepted.
- Numerous studies describing the viability of SARS-CoV-2 on a variety of porous and nonporous surfaces have elucidated the transmission of SARS-CoV-2 through fomite contamination of inanimate surfaces.
- Under experimental conditions, SARS-CoV-2 was found to be more stable on stainless steel and plastic surfaces than copper and cardboard, with viable virus being detected up to 72 hours after inoculation.
- At 20 degrees Celsius, viruses were isolated from nonporous surfaces such as glass and stainless steel for up to 28 days. In contrast, SARS-CoV-2 was recovered less efficiently from porous materials than from nonporous surfaces.
- SARS-CoV-2 can survive on plastic and stainless steel for up to 2-3 days, cardboard for up to 1 day, and copper for up to 4 hours, according to a study evaluating the duration of the virus’s viability on objects and surfaces. Furthermore, it appears that contamination was higher in intensive care units (ICUs) than in general wards, and SARS-CoV-2 can be found on floors, computer mice, trash cans, and sickbed handrails, as well as in the air up to 4 meters from patients, implicating both nosocomial and fomite transmission.
- Recent information from the Centers for Disease Control and Prevention (CDC) indicates that individuals can become infected with SARS-CoV-2 through contact with contaminated surfaces, but the risk is low and this is not the primary mode of transmission.
- Multiple case studies have documented that patients with SARS-CoV-2 infection have live virus in their feces, suggesting the possibility of fecal-oral transmission.
- A meta-analysis involving 936 neonates born to mothers with COVID-19 revealed that vertical transmission is conceivable but rare.
Clinical manifestations of Coronavirus
- fever and dry cough are the most prevalent symptoms.
- Most patients demonstrate bilateral pneumonia.
- SARS-CoV-2 is more likely to affect elderly men with multiple comorbidities.
- Patients’ blood counts reveal leucopenia and lymphopenia.
- ICU patients have higher plasma concentrations of IL2, IL7, IL10, GSCF, IP10, MCP1, MIPA, and TNFa than non-ICU patients.
- There are three levels of severity for COVID-19: moderate, severe, and critical.
- There have been reports of asymptomatic infections, but the majority of asymptomatic patients went on to develop the disease.
- COVID-19 infection can result in myocardial damage, arrhythmic complications, neurological complications, digestive symptoms, liver damage, hypercoagulability, thrombotic complications, and even stroke.
- Critical patients could rapidly develop ARDS, metabolic acidosis that is difficult to correct, septic shock, coagulation dysfunction, and multiple organ functional failure.
- Severe complications include ARDS, RNAaemia, multiple organ failure, and acute cardiac injury.
- About 26.1% of patients were admitted to the ICU due to COVID-19-related complications.
- With appropriate diagnosis and treatment, the majority of COVID-19 patients have a favorable prognosis.
- Patients with underlying diseases and advanced age have a poorer prognosis.
Lab Diagnosis of Coronavirus
The laboratory diagnosis of coronavirus disease (COVID-19) is crucial for confirming the infection, identifying the virus, and taking appropriate measures to prevent further spread. Here are some important points regarding the lab diagnosis of COVID-19:
- Real-time reverse transcription-polymerase chain reaction (RT-PCR) is the most commonly used method for diagnosing COVID-19.
- Samples for RT-PCR testing are typically collected from the respiratory tract, including nasopharyngeal and oropharyngeal swabs, sputum, and bronchoalveolar lavage (BAL) fluid.
- RT-PCR tests detect the viral RNA in patient samples and can provide a rapid diagnosis within a few hours.
- Serological tests, including enzyme-linked immunosorbent assay (ELISA) and chemiluminescent immunoassay (CLIA), are also used to detect COVID-19 antibodies in patient blood samples.
- Serological tests can help identify patients who have previously been infected with COVID-19, but they are not useful for diagnosing acute infections, as it takes several days to weeks for antibodies to develop after infection.
- Rapid antigen tests are available for diagnosing COVID-19, but they have a lower sensitivity compared to RT-PCR and may result in false-negative results.
- Laboratory results should be interpreted in conjunction with clinical symptoms and epidemiological history.
- Laboratory diagnosis of COVID-19 should be performed in biosafety level 2 (BSL-2) or higher laboratories with appropriate safety measures to prevent laboratory-acquired infections.
- Laboratory workers should wear appropriate personal protective equipment (PPE) when handling patient samples and performing diagnostic tests.
- Rapid sharing of laboratory information and test results is essential for effective outbreak response and control.
Treatment of Coronavirus
There is currently no specific treatment for COVID-19, the disease caused by the coronavirus. However, symptomatic treatment and supportive care can improve outcomes for patients. Here are some treatment options and approaches currently being used:
- Antiviral drugs: Some antiviral drugs have shown promise in treating COVID-19, including remdesivir and favipiravir. However, their effectiveness is still being studied.
- Oxygen therapy: For patients with respiratory distress, supplemental oxygen therapy can help maintain oxygen levels in the blood.
- Corticosteroids: For patients with severe cases of COVID-19, corticosteroids may be used to reduce inflammation and improve lung function.
- Supportive care: This includes providing fluids, nutrition, and medication to manage symptoms such as fever and pain.
- Immunomodulatory drugs: Drugs that modulate the immune system, such as tocilizumab, may be used in severe cases to prevent cytokine storms and other immune-related complications.
It is important to note that treatment should be tailored to the individual patient based on their symptoms and disease severity, and that there is currently no single “cure” for COVID-19.
Prevention and control of Coronavirus
Prevention and control of Coronavirus involves measures that aim to reduce the transmission of the virus and protect individuals and communities from infection. Some of the key strategies include:
- Vaccination: COVID-19 vaccines are an effective way to prevent infection and reduce the severity of illness in those who do get infected.
- Personal protective measures: This includes frequent hand washing with soap and water or using alcohol-based hand sanitizers, wearing face masks in public places, and practicing social distancing.
- Testing and contact tracing: Early identification of infected individuals and their close contacts can help to prevent further spread of the virus.
- Quarantine and isolation: People who have been infected or exposed to the virus should isolate or quarantine themselves to prevent further transmission.
- Travel restrictions: Restrictions on travel can help to prevent the spread of the virus between countries or regions.
- Environmental sanitation: Frequent cleaning and disinfection of surfaces and objects can reduce the risk of transmission.
- Health education: Public health campaigns and education can help to promote awareness of the virus, its transmission, and prevention measures.
- Surveillance and monitoring: Monitoring the spread of the virus, detecting outbreaks, and assessing the effectiveness of prevention and control measures can help to guide public health responses.
FAQ
What is Coronavirus?
Coronavirus (COVID-19) is a viral respiratory illness caused by the SARS-CoV-2 virus.
How does Coronavirus spread?
The virus primarily spreads through respiratory droplets when an infected person talks, coughs, or sneezes. It can also spread by touching surfaces contaminated with the virus and then touching your mouth, nose, or eyes.
What are the symptoms of Coronavirus?
Common symptoms include fever, cough, shortness of breath, fatigue, body aches, and loss of smell or taste. Some people may have no symptoms at all.
How is Coronavirus diagnosed?
Coronavirus can be diagnosed through a PCR test that detects the virus’s genetic material in a sample taken from the nose or throat.
How is Coronavirus treated?
Treatment is primarily supportive, with the focus on managing symptoms such as fever and cough. In severe cases, hospitalization may be required, and treatments such as oxygen therapy or mechanical ventilation may be necessary.
How can I protect myself from Coronavirus?
The best ways to protect yourself include wearing a mask, washing your hands frequently, avoiding close contact with others, and staying home if you feel unwell.
Can children get Coronavirus?
Yes, children can get Coronavirus, although they are less likely to have severe symptoms.
Can I get Coronavirus from touching packages or mail?
The risk of getting Coronavirus from touching packages or mail is considered low. However, it is still recommended to wash your hands after handling mail or packages.
Can I get Coronavirus twice?
It is possible to get Coronavirus twice, but it is rare. Some studies suggest that immunity may last for several months to a year.
When will the Coronavirus pandemic end?
It is difficult to predict when the pandemic will end, as it depends on factors such as vaccine distribution and the level of community transmission. However, ongoing efforts such as vaccination campaigns and public health measures are aimed at controlling the spread of the virus.
References
- Li F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu Rev Virol. 2016 Sep 29;3(1):237-261. doi: 10.1146/annurev-virology-110615-042301. Epub 2016 Aug 25. PMID: 27578435; PMCID: PMC5457962.
- Wang, M.-Y., Zhao, R., Gao, L.-J., Gao, X.-F., Wang, D.-P., & Cao, J.-M. (2020). SARS-CoV-2: Structure, Biology, and Structure-Based Therapeutics Development. Frontiers in Cellular and Infection Microbiology, 10. doi:10.3389/fcimb.2020.587269
- https://www.scripps.edu/covid-19/science-simplified/parts-of-a-coronavirus/
- https://web.mit.edu/fnl/volume/324/king_etal.html
- https://www.biophysics.org/blog/coronavirus-structure-vaccine-and-therapy-development
- https://www.nih.gov/news-events/nih-research-matters/novel-coronavirus-structure-reveals-targets-vaccines-treatments
