Table of Contents
What is DNA?
DNA is a polymer of deoxyribonucleotides stuck together (or simply deoxynucleotides). It is made up of deoxyadenylate (dAMP), deoxyguanylate (dGMP), deoxycytidylate (dCMP), and deoxythymidylate (dTMP) units (dTMP) (Some authors prefer to use TMP instead of deoxythymidylate because it is only found in DNA.)
- DNA = deoxy-ribo-nucleic acid
- RNA = ribo-nucleic acid
- deoxy = 2′-H (vs 2′-OH in RNA)
- ribo = ribose sugar = pentose sugar
- acid = phosphate group gives it acidity
- DNA molecules are made up of nucleic acids, which are the molecules in a cell that carry information.
- DNA molecules are made up of many smaller molecules called nucleotides. They are polymers.
- Each nucleotide is made up of a sugar molecule, a phosphate group, and a nitrogenous base.
- The two DNA strands in a DNA molecule are twisted around each other to make a spiral shape called the double helix.
- James Watson, Francis Crick, and Rosalind Franklin were the first people to find out that DNA has a double helix shape in 1953.
- In the nucleus, DNA molecules store the genetic code, which is all the information an organism needs to work.
- In 1953, James Watson, an American biologist, Francis Crick, an English physicist, and Rosalind Franklin were the first people to figure out that DNA has a double helix shape (an English Chemist).
- Even though Watson and Crick were given credit for finding the double helix, it is thought that Franklin’s data helped them find it.
- Franklin was an expert at an imaging method called X-ray crystallography, which she used to make the first picture of the spiral shape of DNA.
- In 1962, the Nobel Prize was given to Watson and Crick for the work they had done. Franklin did help make the discovery, but she did not win the prize because she had died of cancer four years before.
History of DNA
The history of DNA begins with Friedrich Miescher, a Swiss physiological chemist, who made a groundbreaking discovery in 1869. Miescher identified a substance inside the nuclei of human white blood cells, which he called “nuclein.” This substance was later renamed “nucleic acid” and eventually “deoxyribonucleic acid” or “DNA.” Initially, Miescher intended to isolate and study the protein components of white blood cells but stumbled upon a new substance with unique chemical properties. He realized that he had discovered a new type of molecule unlike any protein known at the time.
Miescher’s discovery of nucleic acids went largely unrecognized for over 50 years. In 1919, Russian biochemist Phoebus Levene continued the investigation into the chemical nature of nucleic acids. Levene proposed the “polynucleotide” model based on his studies of yeast nucleic acids. According to his model, nucleic acids were composed of nucleotides, each consisting of a phosphate group, a sugar molecule (ribose in RNA and deoxyribose in DNA), and a nitrogen-containing base. While Levene’s proposed tetranucleotide structure was simplistic and later proven incorrect, he laid the groundwork for understanding the components of nucleic acids.
Erwin Chargaff, an Austrian biochemist, further expanded on Levene’s work. Inspired by Oswald Avery’s discovery that genes are composed of DNA, Chargaff delved into the chemistry of nucleic acids. He investigated the composition of DNA in different species and made two significant observations. Firstly, he found that the nucleotide composition of DNA varies among species, contrary to Levene’s proposed order. Secondly, Chargaff discovered that the amounts of adenine (A) and thymine (T) are usually similar, as are the amounts of guanine (G) and cytosine (C). This discovery, known as “Chargaff’s rule,” was crucial for understanding the pairing of bases within the DNA molecule.
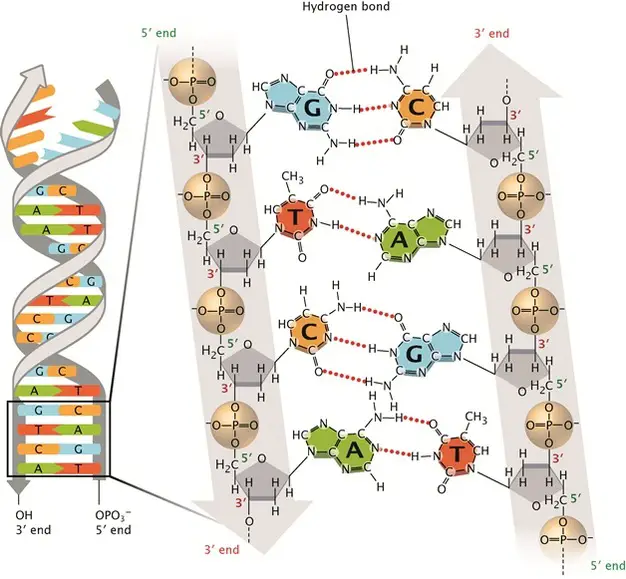
The structure of DNA was eventually unraveled by James Watson and Francis Crick in 1953. Their work was facilitated by advancements in X-ray crystallography by Rosalind Franklin and Maurice Wilkins, as well as the model-building techniques developed by Linus Pauling. Watson and Crick proposed the double helix structure of DNA, with two strands connected by hydrogen bonds. They discovered that A always pairs with T and C always pairs with G, consistent with Chargaff’s rule. Additionally, they determined that the DNA molecule is right-handed and anti-parallel, meaning the two strands run in opposite directions.
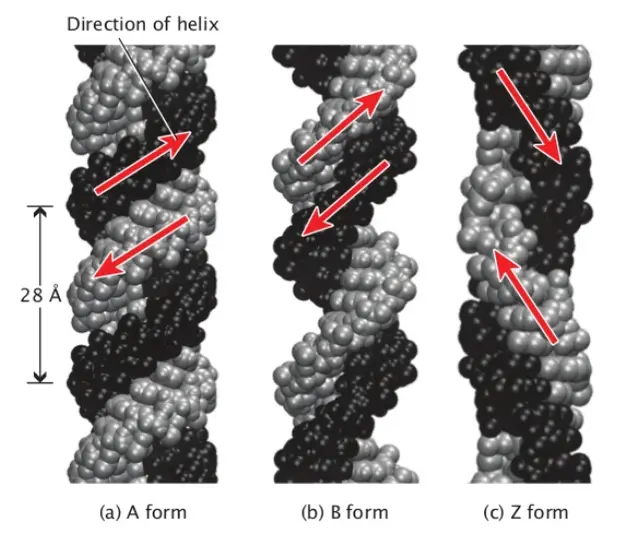
Since Watson and Crick’s model, scientists have made some minor refinements but the four major features of the DNA structure remain unchanged. These features include the double-stranded helix with complementary base pairing, the right-handed helical structure, the anti-parallel arrangement of strands, and the exposed nitrogenous bases available for hydrogen bonding. Scientists have also identified different conformations of the DNA double helix, such as the most common B-DNA, as well as A-DNA and Z-DNA, which occur under specific conditions.
Overall, the history of DNA spans from Friedrich Miescher’s discovery of nucleic acids to Watson and Crick’s proposal of the double helix structure, building upon the contributions of scientists like Levene and Chargaff. This understanding of DNA’s structure and composition laid the foundation for further advancements in genetics and molecular biology.
Properties of DNA
- DNA is a long polymer made up of repeating units called nucleotides. Each nucleotide is usually represented by a single letter: A, T, C, or G.
- Along its length, DNA’s structure changes, so it can twist into tight loops and other shapes.
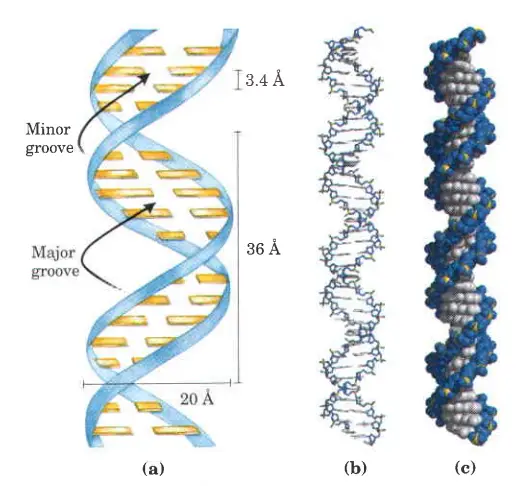
- It is made up of two helical chains that are held together by hydrogen bonds in all species.
- Both chains are wound around the same axis and have the same spacing between links, which is 34 ngstroms (3.4 nm).
- The space between the two chains is 10. (1.0 nm).
- When the DNA chain was measured in a different solution, it was 22–26 (2.2–2.6 nm) wide, and one nucleotide unit was 3.3 (0.33 nm) long.
- Even though each nucleotide is very small, a DNA polymer can be very long and may have hundreds of millions of nucleotides, as in chromosome 1. The largest human chromosome is chromosome 1. It has about 220 million base pairs and would be 85 mm long if it were straight.
- DNA is usually made up of two strands that are held together tightly.
- In the shape of a double helix, these two long strands wrap around each other.
- The nucleotide contains a portion of the molecule’s backbone (which holds the chain together) and a nucleobase (which interacts with the other DNA strand in the helix).
- A nucleobase that is connected to a sugar is called a nucleoside. A nucleotide is a base that is connected to a sugar and one or more phosphate groups.
- A polynucleotide is a biopolymer made up of many linked nucleotides, like DNA.
- The backbone of a strand of DNA is made up of groups of phosphate and sugar that come and go.
- 2-Deoxyribose is the sugar in DNA. It is a pentose sugar, which means it has five carbons.
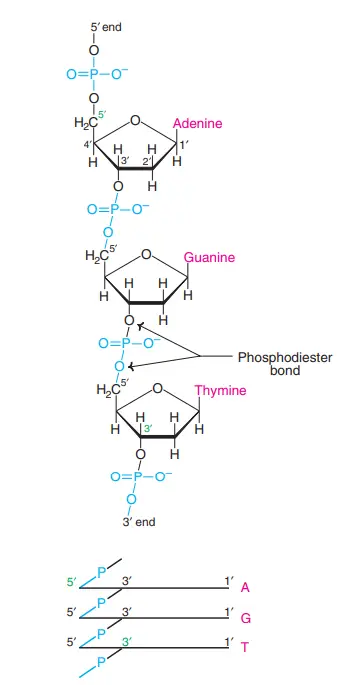
- The sugars are held together by phosphate groups that forge phosphodiester connections between the third and fifth carbon atoms of adjacent sugar rings. These are the 3′-end (three prime end) and 5′-end (five prime end) carbons. The prime symbol is used to distinguish these carbon atoms from those of the base with which the deoxyribose forms a glycosidic bond.
- So, a DNA strand usually has one end with a phosphate group attached to the 5′ carbon of a ribose (5′ phosphoryl) and the other end with a free hydroxyl group attached to the 3′ carbon of a ribose (3′ hydroxyl).
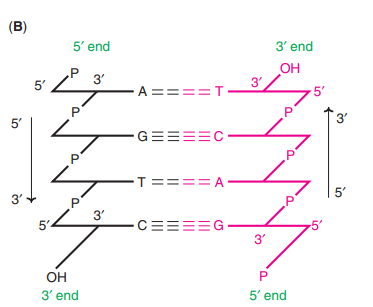
- Each DNA strand has directionality (sometimes called polarity) because of how the 3′ and 5′ carbons line up along the sugar-phosphate backbone.
- In a double helix of nucleic acid, the direction of the nucleotides in one strand is the opposite of the direction of the nucleotides in the other strand. This means that the two strands are antiparallel.
- The asymmetric ends of DNA strands are called five prime ends (5′) and three prime ends (3′). The 5′ end has a phosphate group at the end, while the 3′ end has a hydroxyl group at the end.
- The sugar is a big difference between DNA and RNA. DNA has 2-deoxyribose, but RNA has ribose, which is a related pentose sugar.
- The DNA double helix is mostly held together by two forces: hydrogen bonds between nucleotides and base-stacking interactions between aromatic nucleobases.
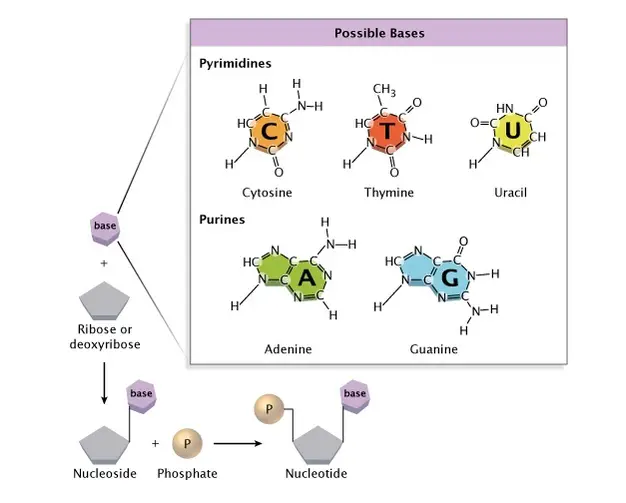
- Adenine (A), cytosine (C), guanine (G), and thymine (T) are the four bases in DNA (T).
- As shown for adenosine monophosphate, these four bases are joined to the sugar-phosphate to make the full nucleotide.
- A-T base pairs are made when adenine pairs with thymine and guanine pairs with cytosine.
- There are two types of nucleobases: the purines, A and G, which are fused heterocyclic compounds with five or six rings, and the pyrimidines, C and T, which are also six-ringed compounds.
- Uracil (U), a fifth pyrimidine nucleobase, usually takes the place of thymine in RNA. It is different from thymine because its ring does not have a methyl group. In addition to RNA and DNA, scientists have made many artificial versions of nucleic acids to study their properties or to use in biotechnology.
- DNA has bases that have been changed. 5-methylcytosine was the first of these to be found. It was found in the genome of Mycobacterium tuberculosis in 1925. The reason that bacterial viruses (bacteriophages) have these noncanonical bases is to avoid the restriction enzymes that are found in bacteria. At least in part, this enzyme system works as a molecular immune system to protect bacteria from getting sick from viruses. The most common DNA bases that can be changed, cytosine and adenine, play important roles in how plants and animals control the expression of their genes.
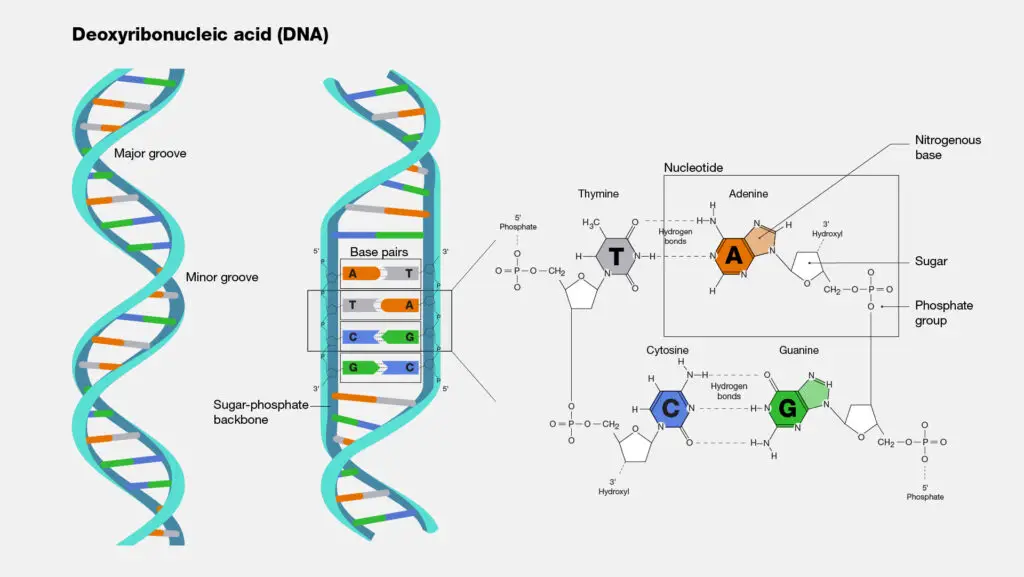
- The backbone of DNA is made up of two helical strands. By following the grooves between the strands, you can find another double helix. These empty spaces are close to the base pairs and may serve as a place to bind. Since the strands are not all lined up the same way, the grooves are not all the same size. The width of the major groove is 22 (2.2 nm), while the width of the minor groove is 12 (1.2 nm).
- The edges of the bases are easier to reach in the major groove than in the minor groove because the major groove is wider. Therefore, proteins like as transcription factors that can bind to particular sequences in double-stranded DNA typically establish contact with the exposed bases in the main groove. This happens in different ways depending on how the DNA is twisted inside the cell, but the major and minor grooves are always named after the different widths that would be seen if the DNA was untwisted and put back into its normal B form.
- A process called “DNA supercoiling” can twist DNA like a rope.
- DNA can take many different shapes, such as A-DNA, B-DNA, and Z-DNA, but only B-DNA and Z-DNA have been directly seen in living organisms. DNA’s shape depends on its level of hydration, its sequence, the amount and direction of supercoiling, chemical changes to the bases, the type and amount of metal ions, and the presence or absence of polyamines in solution.
- Because DNA has phosphate groups, it is acidic like phosphoric acid and can be thought of as a strong acid. At a normal pH level in the cell, it will be fully ionised, releasing protons that leave negative charges on the phosphate groups. By repulsion, nucleophiles that could break down DNA by hydrolysis are pushed away by the negative charges.
- Several artificial nucleobases have been made, and they work well in Hachimoji DNA, which is an eight-base version of DNA. These artificial bases, which have been given the names S, B, P, and Z, can bond in a predictable way (S to B and P to Z), keep the double helix shape of DNA, and be turned into RNA.
- Fraying happens in DNA when there are parts that aren’t complementary at the end of a double-strand of DNA that is otherwise complementary. But branched DNA can happen if a third strand of DNA is added and has parts that can connect to the broken parts of the double-strand DNA that is already there. Even though three strands of DNA make up the simplest example of branched DNA, complexes with more strands and more branches are also possible. [63] In nanotechnology, branched DNA can be used to make geometric shapes. See the section on how it is used in technology below for more information.
- If a DNA sequence is the same as a copy of messenger RNA that is turned into protein, this is called a “sense” sequence. “Antisense” refers to the sequence on the other strand of DNA. On different parts of the same strand of DNA, there can be both sense and antisense sequences (i.e. both strands can contain both sense and antisense sequences).
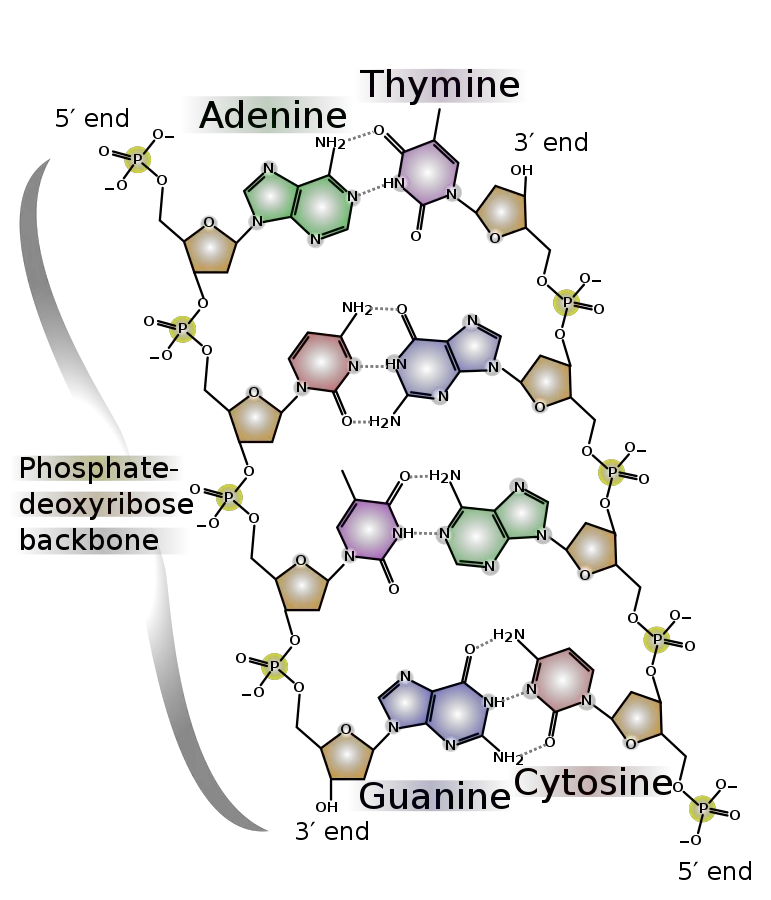
Structure of DNA
- DNA is a polymer composed of deoxyribonucleotides joined by phosphodiester linkages.
- It contains adenine, guanine, cytosine, and thymine as bases.
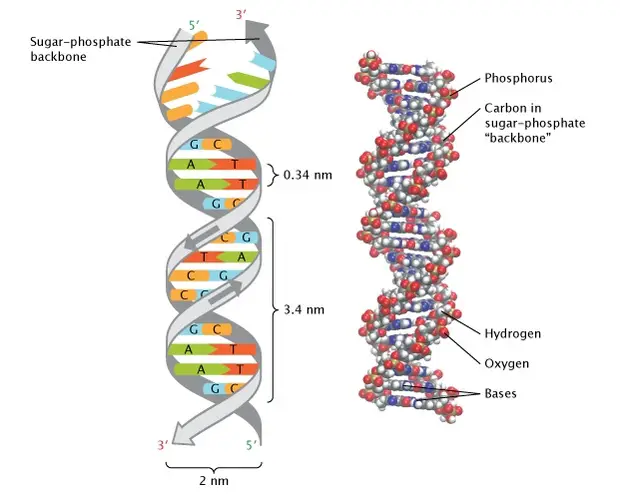
- DNA molecules are typically made of two polynucleotide chains coiled together to create a 2.0 nm-diameter double helix.
- The monomers of DNA are referred to as deoxyribonucleotides due to the presence of deoxyribose inside them.
- Because it consists of a phosphate that forms a bridge between the 3′-hydroxyl of one sugar and the 5′-hydroxyl of an adjacent sugar, the connection that connects the monomers to form the polymer is known as a phosphodiester bond.
- Attached to the 1′-carbon of the deoxyribose sugars are the purine and pyrimidine bases, which extend toward the centre of the cylinder formed by the two chains. (The numbers denoting the carbons in sugars are prefixed with a prime to differentiate them from the numbers denoting the carbons and nitrogens in nitrogenous bases.)
- Each strand’s bases interact with those of the other strand to produce base pairs.
- One base pair is layered on top of the next every 0.34 nm at the centre of the structure.
- Two hydrogen bonds always connect the purine adenine (A) of one strand to the pyrimidine thymine (T) of the opposing strand.
- The purine guanine (G) forms three hydrogen bonds with cytosine (C).
- Due to the pairing of AT and GC bases, the two strands of a DNA double helix are complementary. In other words, the bases in one strand correspond to those in the other strand according to precise principles for base pairing.
- Due to the fact that the sequences of bases in these strands encode genetic information, much work has gone into identifying the base sequences of DNA and RNA from several species, including hundreds of bacteria.
- The two DNA polynucleotide strands join together similarly to jigsaw puzzle pieces.
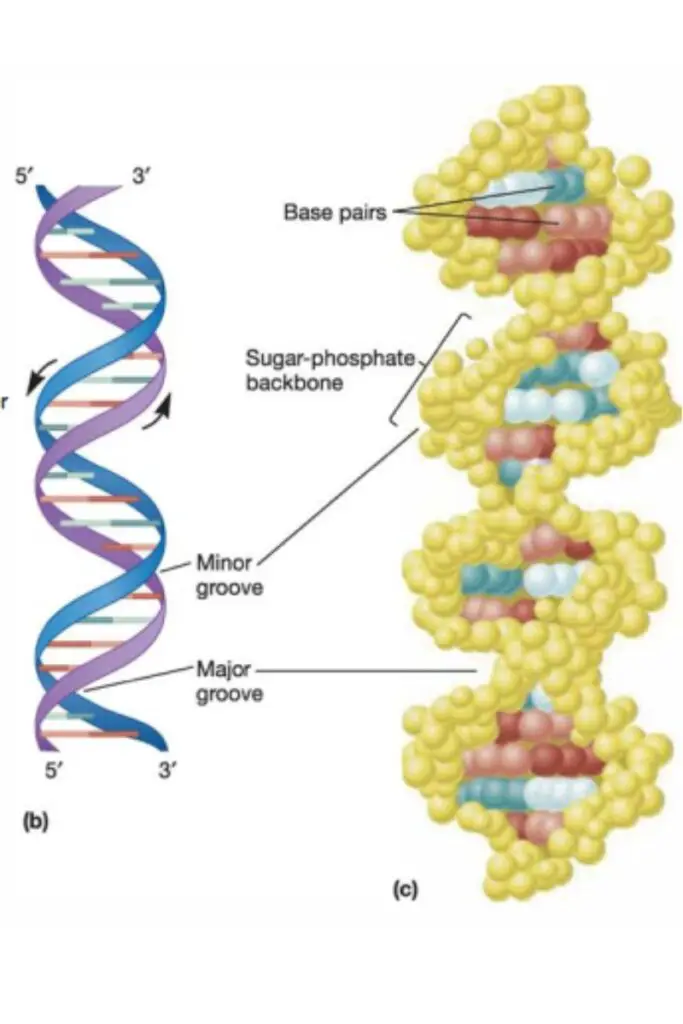
- The two strands are not parallel to one another. When the strands twist around one another, the backbone forms a wide major groove and a narrower minor groove.
- There are 10.5 base pairs per turn of the helix, and the vertical length of each turn is 3.4 A.
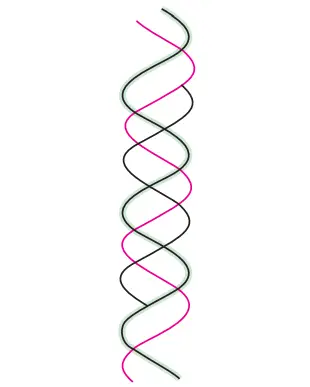
- The helix is right-handed, meaning that the chains rotate counterclockwise as they approach an observer observing the longitudinal axis.
- The two backbones are antiparallel with respect to the orientation of their sugars, meaning they run in opposite directions.
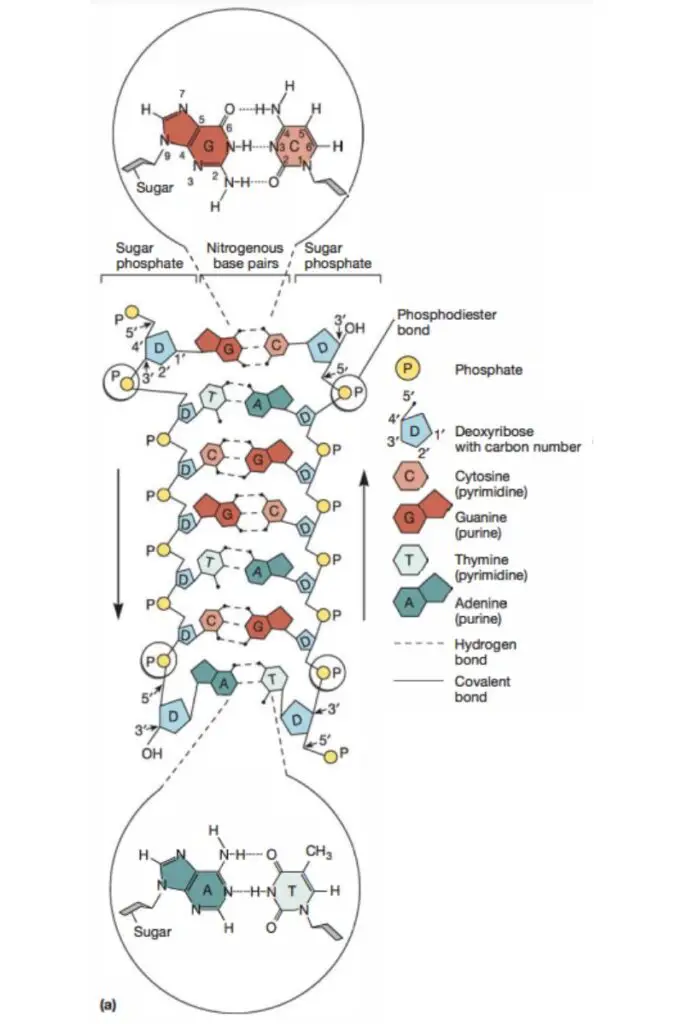
- One end of each strand has an exposed 5′-hydroxyl group, frequently with connected phosphates, whereas the other end has a free 3′-hydroxyl group.
- One strand is orientated 5′ to 3′ and the other, 3′ to 5′, in a specified direction.
- The DNA structure just described corresponds to the B form, the most prevalent type in cells.
- Two additional kinds of DNA have been discovered. The primary difference between the A form and the B form is that the A form has 11 base pairs per helical turn, as opposed to 10.5, and a vertical length of 2.6 A, as opposed to 3.4 A. It is therefore wider than the B variant.
- The Z form has a left-handed helical structure, as opposed to the right-handed helical structure of the B and A forms.
- The Z shape possesses 12 base pairs per helical revolution and 3.7 A of vertical rise. Therefore, it is slimmer than the B variant. It is still unknown whether the A form is present in cells.
- Nevertheless, there is evidence that tiny sections of chromosomes can exist in the Z form. Unknown is the function, if any, of these sections of Z DNA.
- Supercoiling is another DNA feature that needs to be addressed. DNA is helical, i.e., it is coiled. When the rotation of a coil is impeded in any way, the coil will coil upon itself. Supercoiling refers to the coiling of a coil.
- Remember that the majority of bacterial chromosomes are closed double-stranded DNA molecules. In this position, the two strands cannot rotate freely with respect to one another, and the molecule is said to be under tension. Supercoiling relieves the tension.
- Positive and negative supercoiling are the two types. Changes in the number of base pairs per turn in the double helix characterise these for DNA.
- As previously mentioned, the B type of DNA has 10.5 base pairs per helix turn.
- Negative supercoiling is supercoiling in which the number of base pairs each turn decreases.
- Positive supercoiling refers to supercoiling that increases the number of base pairs every turn.
- Generally, bacterial chromosomes are negatively supercoiled.
What is the importance of supercoiling?
- DNA is compacted by supercoiling so that it can fit within the cell.
- Additionally, supercoiling “loosens” the DNA, making it easier to separate the two strands.
- In both DNA replication and transcription, separation of the two strands is a crucial early step.
- Additionally, positive supercoiling is frequently introduced during DNA replication. This can inhibit DNA replication, so it must be eliminated.
The Size Of Dna Molecule —Units Of Length
DNA molecules are enormous. A pair of B-DNA with a thickness of 0.34 nm has an average molecular mass of 660 daltons.
For length measurement, the DNA double-stranded structure is taken into account and expressed in terms of base pairs (bp). A kilobase pair (kb) is 103 base pairs (bp), while a megabase pair (Mb) is 106 base pairs (bp), and a gigabase pair (Gb) is 109 base pairs (bp). The relationships between kb, Mb, and Gb can be summarised as follows:
- 1 kb = 1000 bp
- 1 Mb = 1000 kb = 1,000,000 bp
- 1 Gb = 1000 Mb = 1,000,000,000 bp
Since most RNAs are single-stranded, the lengths of RNA molecules (like DNA molecules) cannot be stated in base pairs (bp).
The length of DNA varies between species and is typically expressed in terms of composition of base pairs and contour length. The length of the contour shows the total length of genomic DNA in a cell. Some species with bp and contour lengths are provided as examples.
- λ phage virus — 4.8 u 104 bp — contour length 16.5 Pm.
- E. coli — 4.6 u 106 bp — contour length 1.5 Pm.
- Diploid human cell (46 chromosomes) — 6.0 u 109 bp — contour length 2 meters.
It should be emphasised that genomic DNA is typically significantly larger than the cell or nucleus it resides in. In humans, a 2-meter-long DNA is packed into a nucleus with a diameter of approximately 10 um.
DNA can occur in either linear or circular forms. The majority of bacterial DNAs exist as closed circles. This contains the DNA of bacterial chromosomes and the DNA of plasmids that is extrachromosomal. Eukaryotic mitochondria and chloroplasts also contain circular DNA.
The majority of chromosomal DNAs in higher organisms are linear. Individual human chromosomes are comprised of a single DNA molecule of varied size. Therefore, the smallest chromosome comprises 34 megabytes while the largest contains 263 megabytes.
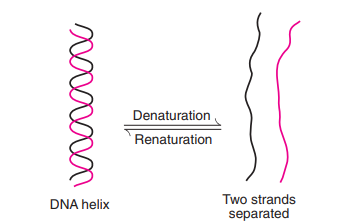
Denaturation Of Dna Strands
Denaturation
- The two DNA strands are kept together by hydrogen bonds. Separation of polynucleotide strands comes from the disruption of hydrogen bonds (either by a change in pH or an increase in temperature).
- This process of DNA losing its helical form is known as denaturation.
- By denaturation, the phosphodiester linkages are not broken. Increase in absorbance at 260 nm can measure the loss of helical structure (in a spectrophotometer).
Hyperchromicity
- Following denaturation, a rise in the absorbance of purines and pyrimidines is referred to as hyperchromicity.
Melting temperature
- The melting temperature (Tm) is the temperature at which DNA loses half of its helical helix.
- Since G-C base pairs are more stable (due to three hydrogen bonds) than A-T base pairs (due to two hydrogen bonds), DNAs with a higher G-C composition have a bigger Tm.
- Therefore, the Tm is 65°C for a G-C level of 35% and 70°C for a G-C value of 50%.
- Formamide destabilises hydrogen bonds of base pairs and, consequently, reduces Tm.
- This chemical substance is utilised successfully in DNA recombination research.
Renaturation
- Renaturation (or reannealing) is the process whereby split complementary DNA strands can reassemble into a double helix.
Organization Of Dna In The Cell
- As previously noted, the length of the double-stranded DNA helix in each chromosome is thousands of times the diameter of the nucleus.
- In humans, a 2-meter-long DNA is packed into a nucleus of 10 um in diameter!
- This is made possible by the incredible packaging and organising of DNA within the cell.
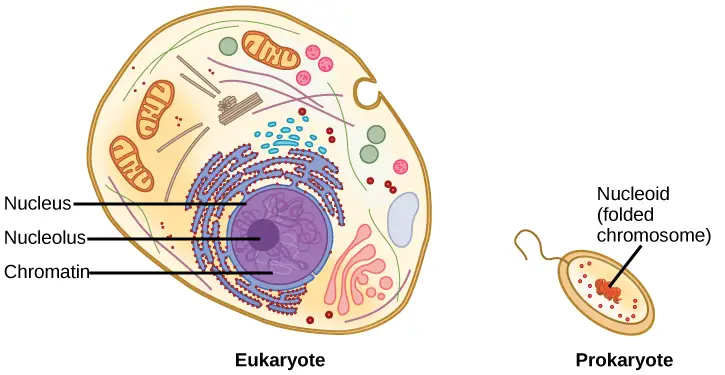
Organization of prokaryotic DNA
- DNA is structured into a single chromosome in the form of a double-stranded circle in prokaryotic cells.
- By interacting with proteins and certain cations, these bacterial chromosomes are packed in the form of nucleoids (polyamines).
Organization of eukaryotic DNA
- In eukaryotic cells, DNA is coupled with numerous proteins to form chromatin, which is then ordered into chromosomes, which are compact structures.
- The DNA double helix is encircled by core proteins, notably histones, which are fundamental in nature.
- The core is made of two histone molecules (H2A, H2B, H3 and H4).
- The fundamental unit of chromatin is the nucleosome, which consists of a core surrounded by two turns of DNA (about 150 bp).
- Separating nucleosomes is spacer DNA to which histone H1 is linked.
- This continuous string of nucleosomes, which represents the beads-on-a-string type of chromatin, is known as a 10-nm fibre.
- The production of 10 nm fibre significantly reduces the length of the DNA.
- This 10-nm fibre is subsequently coiled to create a 30-nm fibre with a solenoid structure that contains six nucleosomes each turn.
- These 30-nm fibres are structured into loops by anchoring A/T-rich scaffold-associated regions (SARS) to a protein scaffold.
- During mitosis, the loops become more tightly wound, and the chromosomes condense and become visible.
Component of DNA
- DNA consists of two strands of nucleotides that repeat. Each nucleotide has three constituents. These elements include:
- Phosphate Group
- 2-deoxyribose sugar
- A nitrogen containing base
- cytosine
- adenine
- Guanine
- thymine
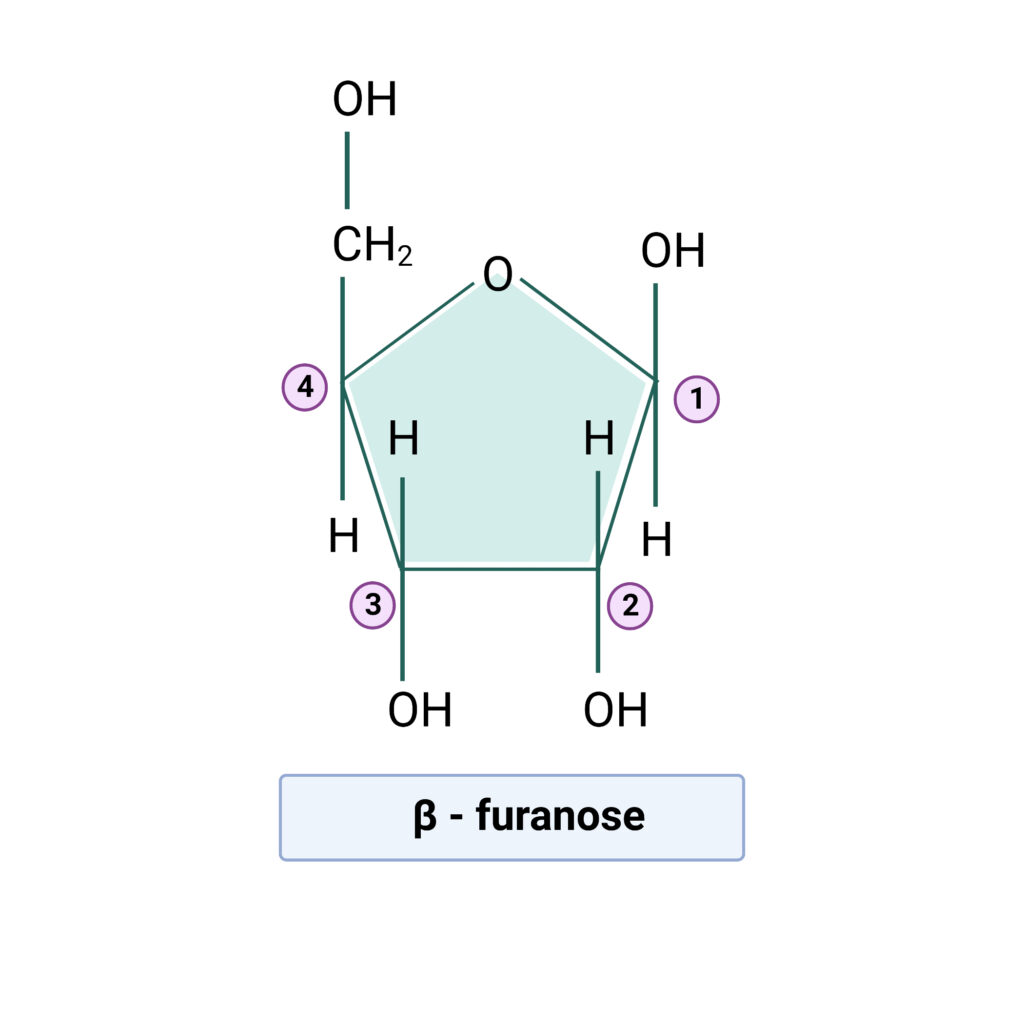
Ribose
- The pentose sugar ribose is one of the basic components of DNA.
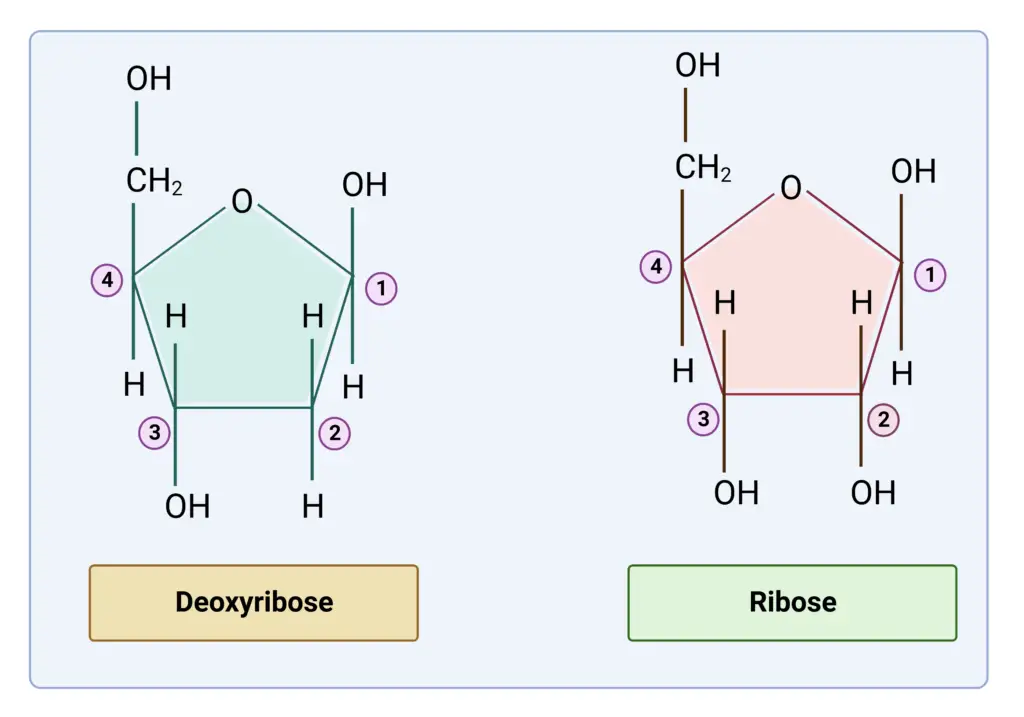
- Both kinds of nucleic acid, DNA and RNA, are composed of the sugar pentose. DNA is composed of deoxyribose and 2′-deoxy D- ribose, whereas RNA is composed of ribose or D-ribose alone.
- Both forms of ribose found in nucleic acid contain five-carbon pentose and beta- furanose ( in a cyclic manner).
- The structure of beta-furanose is shown in the diagram below.
The RNA has Uracil instead of thymine, however this is insufficient to differentiate it from DNA. Even if some uracil is present in DNA, 2′-deoxy D-ribose transforms it into DNA. Similarly, even if thymine is present in RNA, D-ribose transforms it into RNA. The difference in the pentose sugar determines the identity of each nucleic acid, not the bases.
Phosphate
- The backbone of nucleic acid (DNA) is composed of phosphate and is commonly referred to as triphosphate since it consists of three phosphate molecules.
- The monophosphate, diphosphate, and triphosphate each contain one, two, and three phosphate units. DNA is referred to as deoxyribonucleotide triphosphate because it contains three phosphates.
- If a single phosphate is present, the compound is known as deoxyribonucleotide monophosphate; if two phosphates are present, the compound is known as deoxyribonucleotide diphosphate.
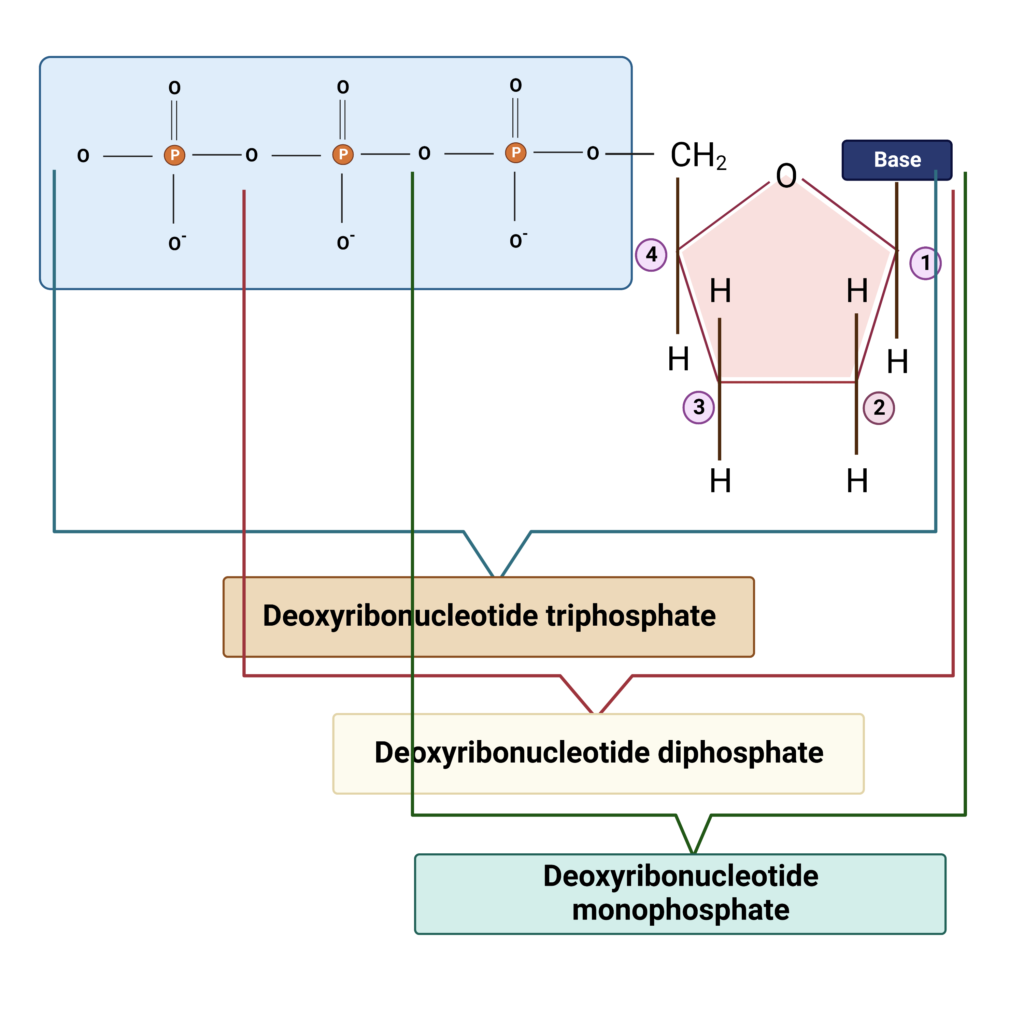
Nitrogenous bases
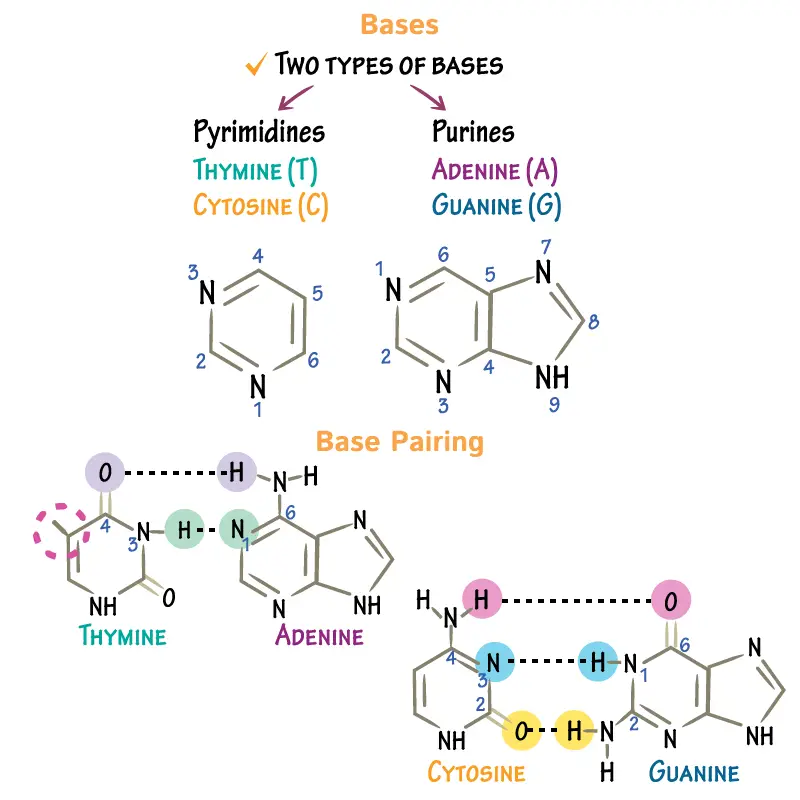
- DNA contains four distinct nitrogenous bases: adenine, thymine, cytosine, and guanine. In RNA, Uracil is present instead of thymine.
- There are two broad categories of nitrogenous bases: purines and pyrimidines.
- Adenine and Guanine are the purines, while Cytosine, Thymine, and Uracil are the pyrimidines.
- Purines and pyrimidines are heterocyclic aromatic chemicals. Both are insoluble in water at a neutral pH, but their solubility increases as the pH decreases. The nature of purines and pyrimidines is hydrophobic.
- The thymine was originally recovered from the tissues of the thymus, hence its name, and the guanine was extracted from bird guano.
- pyrimidines are composed of a six-membered carbon-nitrogen ring with two nitrogen atoms, whereas purines are composed of a six-membered and five-membered-nitrogen containing carbon-nitrogen ring with four nitrogen atoms.
Nomenclature
Purines:
- Guanine: 2-amino-6-oxy purine
- Adenine: 6-amino purine
Pyrimidines:
- Thymine: 2,4-dioxy-5-methyl pyrimidine
- Uracil: 2,4-dioxy pyrimidine
- Cytosine: 2-oxy-4-amino pyrimidine
In addition to xanthine and hypoxanthine, nature also contains orotic acid-pyrimidines, which are not contained in DNA or RNA.
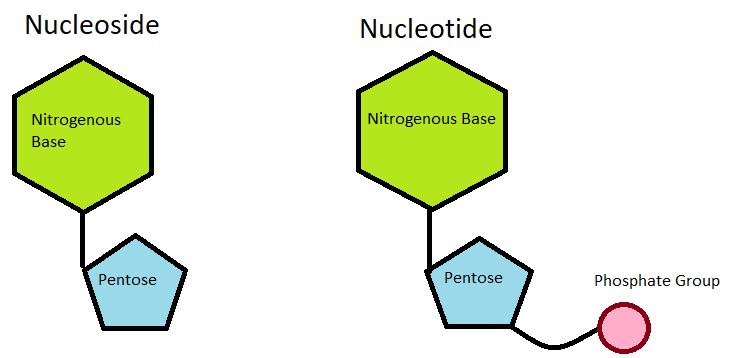
Nucleotide
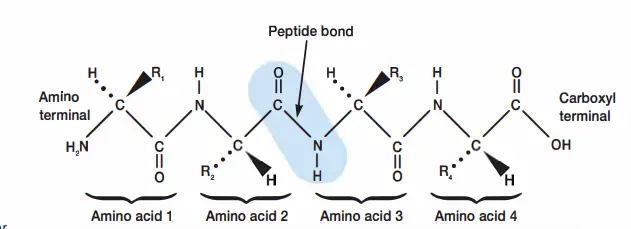
- A nucleotide comprises pentose sugar, a nitrogen-containing base (nitrogenous base), and phosphate.
- A nucleotide is a structural component of DNA. Each nucleotide forms a hydrogen link with the nucleotide of another strand and a phosphodiester bond with neighbouring nucleotides.
- By forming hydrogen bonds and phosphodiester bonds, the nucleotides give DNA its helical shape. A nucleotide lacking phosphate is referred to as a nucleoside (deoxyribonucleoside).
- There are also methylated nucleotides in nature, which assist activate and deactivate gene expression and safeguard genetic information.
Bonds in DNA
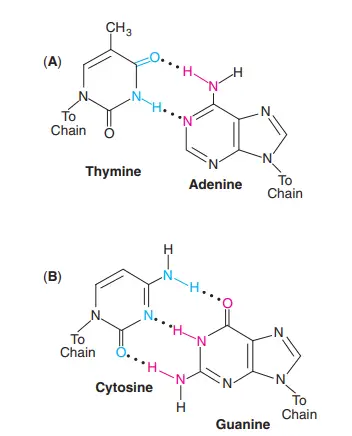
- A hydrogen bond formed between the nitrogenous bases of the two strands of DNA.
- Similar to the electronegative atoms nitrogen and oxygen, a weak chemical link known as a hydrogen bond formed. The hydrogen bond is a transient binding that occurs exclusively at shorter distances and stabilises DNA.
- Some chemical treatments and heat can readily cause its decomposition.
- Two strands of DNA are connected by three weak hydrogen bonds between Guanine and Cytosine and two weak hydrogen bonds between Adenine and Thymine.
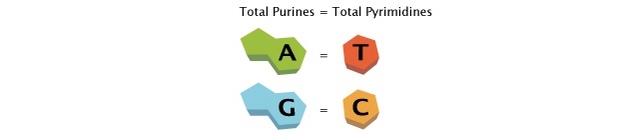
- According to the Watson and Crick model, the DNA double helix follows chargeoff laws. The number of G and A in the DNA is equivalent to the number of C and T.
- Hydrogen bonds are the fundamental components of the polymerase chain reaction. As the number of hydrogen bonds grows, so does the temperature of denaturation.
- Furthermore, the denaturation time increases as the overall GC concentration rises. In general, 95 °C and 5 minutes of denaturation are sufficient to sever all hydrogen bonds between double-stranded DNA.
- UV radiation with a wavelength of 260nm is absorbed by DNA. DNA with a single strand absorbs more UV radiation than DNA with two strands.
- In addition, weak Van der Waals forces, Base-stacking interactions, and a mild hydrophobic effect serve to stabilise the DNA by acting between the bases.
Phosphodiester bond
- The 3′ carbon of one deoxyribose sugar forms a phosphodiester link with the 5′ carbon of the second deoxyribose sugar. The phosphodiester bond forms the DNA’s backbone. After the pyrophosphate is liberated, the bond is created.
- The 5′ or 3′ position without a nucleotide is considered the “end” of DNA. The end of DNA containing a free phosphate is known as the 5′ end, whereas the end containing a free hydroxyl group is known as the 3′ end.
- In addition to the phosphodiester bond and the hydrogen bond, the glycosidic bond is an essential bond. The link between the pentose sugar and bases was glycosidic. All of the linkages create a stable, more electronegative double-helix structure in DNA.
Types of DNA
- B-DNA: the typical double helix structure of double-stranded DNA; see diagram at deoxyribonucleic acid.
- Z-DNA: Z-DNA is a type of DNA in which the phosphate groups form a dinucleotide repeating unit zigzagging along a left-handed helix with a single, deep groove; it is more likely to occur in stretches of alternating purines and pyrimidines.
- A-DNA: Initial identification was based on X-ray diffraction study of DNA fibres at 75% relative humidity.
- D-DNA: A rare variation having eight base pairs per helical turn and no guanine in its structure.
- E- DNA: Extended or unusual DNA.
- Spacer DNA: the nucleotide sequences that occur between genes; in eukaryotes, these sequences are frequently lengthy and comprise several repeating sequences; in particular, the DNA that occurs between genes encoding ribosomal RNA.
- Complementary or copy DNA (cDNA): synthetic DNA transcribed from a specific RNA via the reverse transcriptase enzyme process.
- Nuclear DNA (nDNA): The DNA of chromosomes found in the nucleus of eukaryotic cells.
- Mitochondrial DNA (mtDNA): the DNA of the mitochondrial chromosome, which is inherited solely from the mother and exists in several thousand copies per cell. Its code is distinct from that of nuclear DNA and any living prokaryote, and it evolves five to ten times faster than nuclear DNA.
- Recombinant DNA: Recombinant DNA is a DNA molecule made of linked sequences that do not ordinarily occur in the same molecule, such as a bacterial plasmid into which a length of viral DNA has been inserted.
- Single copy DNA (scDNA): The bulk of gene sequences encoding polypeptides in eukaryotes are single copy DNA (scDNA).
- Repetitive DNA: Repetitive DNA consists of nucleotide sequences that occur several times within a genome; they are characteristic of eukaryotes and do not typically encode polypeptides. Clustered or dispersed sequences may be moderately (10 to 104 copies per genome) or extensively (>106 copies per genome) repetitive. Some structural genes for ribosomal RNA and histones are encoded by moderately repetitive DNA sequences; the majority of highly repetitive sequences are satellite DNA.
| Form | Direction | Bases/ 360o Turn | Helix Diameter |
|---|---|---|---|
| A | Right | 11.0 | 23A |
| B | Right | 10.0 | 19A |
| C | Right | 9.3 | 19A |
| Z | Left | 12.0 | 18A |
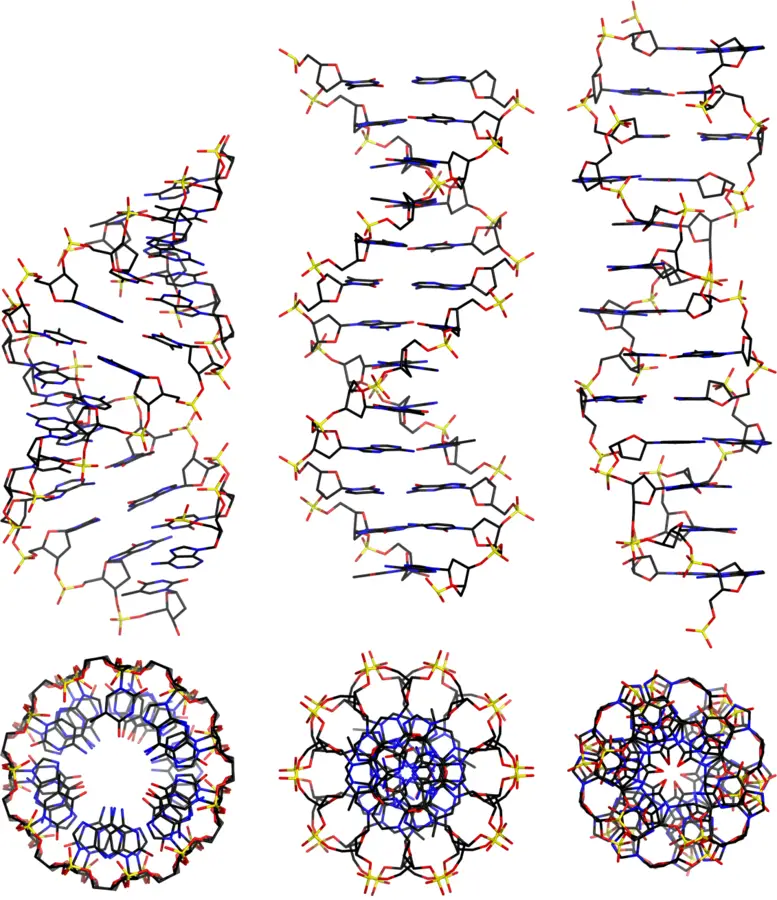
Other Types of DNA
Bent DNA
- In general, DNA sequences containing the adenine base are stiff and straight.
- When A-tracts are replaced by other bases or when the helix collapses into the minor groove of A-tract, DNA has a curved shape.
- DNA structural bending has also been attributed to photochemical damage or mispairing of nucleotides.
- Certain anticancer medications (e.g. cisplatin) induce bent structure in DNA. This altered structure can absorb proteins that harm DNA.
Triple-stranded DNA
- The creation of triple-stranded DNA may result from extra hydrogen bonds between the nucleotides.
- Thus, a thymine can make two Hoogsteen hydrogen bonds with the adenine in an A-T pair to generate T-A-T.
- Similarly, a protonated cytosine can establish two hydrogen bonds with guanine of G–C pairings, resulting in the formation of C–G–C.
- The triple helix is less stable than the double helix. This is because the triple helix’s three negatively charged backbone strands result in a greater electrostatic repulsion.
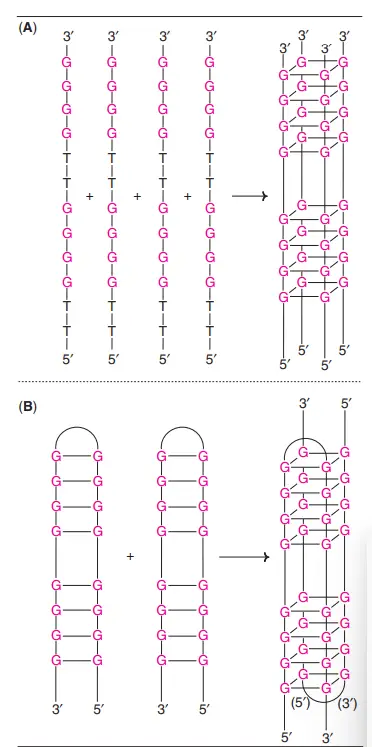
Four-stranded DNA
- Extremely guanine-rich polynucleotides can create a unique tetrameric structure known as G-quartets.
- Hoogsteen hydrogen bonds hold these planar structures together.
- G-tetraplexes, antiparallel four-stranded DNA structures, have also been found.
- The telomeres at the ends of eukaryotic chromosomes, which are rich in guanine, create G-tetraplexes.
- In recent years, anti-cancer chemotherapies have focused on telomeres.
- G-tetraplexes have been linked to the recombination of immunoglobulin genes and the dimerization of human immunodeficiency virus double-stranded genomic RNA (HIV).
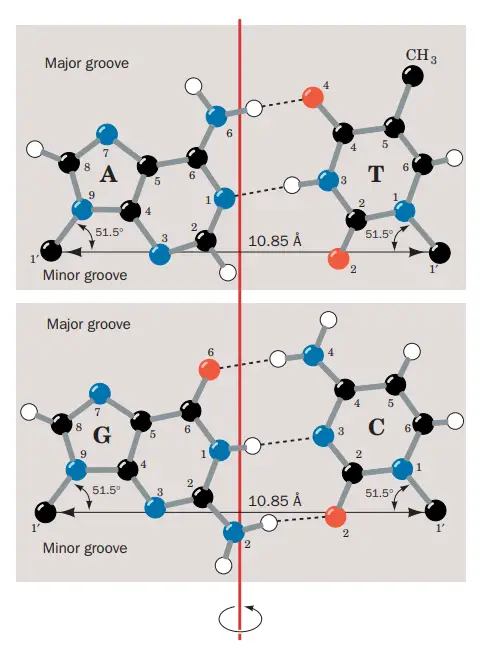
What are Major and Minor Grooves of the DNA?
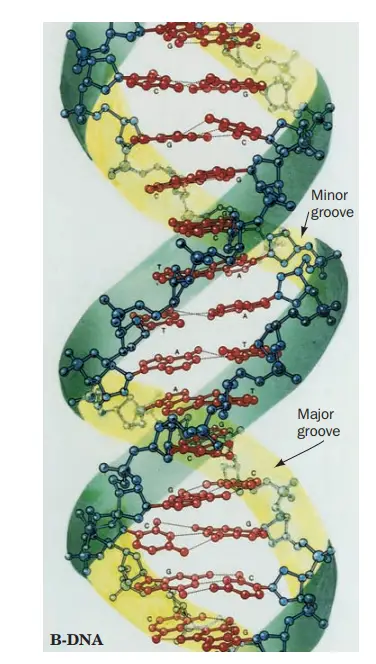
- Due to DNA’s double helix structure, the molecule has two asymmetric grooves. One groove is more diminutive than the other.
- This asymmetry is caused by the geometrical structure of the bonds between the phosphate, sugar, and base groups, which forces the base groups to bind at 120-degree angles as opposed to 180-degree angles.
- The wider groove, known as the major groove, happens when the backbones are widely separated, whereas the smaller groove, known as the minor groove, occurs when the backbones are close together.
- Due to the fact that the major and minor grooves show the margins of the bases, they can be utilised to determine the base sequence of a particular DNA molecule.
- Proteins must be able to recognise certain DNA sequences to which they can bind in order for the body and cell to carry out their respective duties.
DNA is a Double Helix Experiment – Watson and Crick model of DNA
In 1953, James Watson and Francis Crick postulated the double helix structure of DNA (Nobel Prize, 1962). The elucidation of DNA structure is regarded as a watershed moment in the history of contemporary biology. The structure of the DNA double helix resembles a ladder that has been bent. The distinguishing characteristics of the Watson-Crick model of DNA (now known as B-DNA) are outlined below.
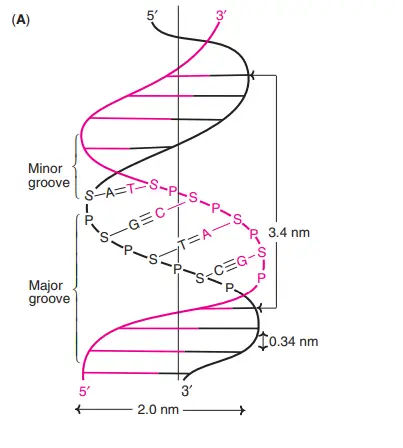
- The DNA is a double right-handed helix. It consists of two polydeoxyribonucleotide chains (strands) that are twisted around a common axis.
- The two strands are antiparallel, meaning that one strand runs from 5c to 3c and the other from 3c to 5c. This is analogous to two neighbouring parallel streets carrying traffic in opposite directions.
- 20 A° is the width (or diameter) of a double helix (2 nm).
- Each turn (pitch) of the helix is 34 A° (3.4 nm), and 10 pairs of nucleotides are spaced around 3.4 A° apart.
- Each strand of DNA has a hydrophilic deoxyribose phosphate backbone (3c-5c phosphodiester linkages) on the exterior (periphery) of the molecule, while the hydrophobic bases are stacked on the interior (core).
- Due to base pairing, the two polynucleotide chains are not identical but complementary.
- The two strands are linked together by complimentary base pairs that generate hydrogen bonds. The A-T pair has two hydrogen bonds, whereas the G-C pair has three. The G C is approximately 50% stronger than the A = T.
- The only two molecules that create hydrogen bonds are purine and pyrimidine. If two purines are facing one other, they cannot occupy the allotted space. And it would be impossible for two pyrimidines to establish hydrogen bonds. A-T, T-A, G-C, and C-G is the only feasible base arrangement in the structure of DNA based on spatial considerations.
- Chargaff’s rule is demonstrated by the complementary base pairing in the DNA helix. Adenine contains the same amount of thymine as thymine, and guanine contains the same amount of cytosine as cytosine.
- The genetic information is located on either the template strand or the sense strand. Antisense strand is the opposing strand. Along the phosphodiester backbone, the double helix features (broad) major grooves and (narrow) minor grooves. At these grooves, proteins interact with DNA without breaking the base pairs or double helix.
DNA is a Double Helix That Stores Genetic Information – Experiments
- Friedrich Miescher initially isolated and described DNA in 1868. He called the material containing phosphorus nuclein.
- The work of Oswald T. Avery, Colin Macleod, and Maclyn McCarty in the 1940s provided the first persuasive proof that DNA was the genetic material.
- Avery and his colleagues discovered that injecting DNA from a virulent strain of Streptococcus pneumoniae into a nonvirulent strain of the same bacteria converted the nonvirulent strain into a virulent strain.
- They determined that the virulent strain’s DNA included the genetic information for virulence.
- Then, in 19b2, tests conducted by Alfred D. Hershey and Martha Chase on the invasion of bacterial cells by a virus (bacteriophage) containing radioactively labelled DNA or protein eliminated any remaining question that DNA, and not protein, contained genetic information.
What is Chargaff’s Rule?
The work of Erwin Chargaff and his colleagues in the late 1940s provided an additional key hint to the DNA structure. They discovered that the four nucleotide bases of DNA are present in varying proportions in the DNAs of various creatures and that the proportions of specific bases are closely connected. Ied Chargaff drew the following conclusions from the DNAs of a great number of different species obtained as part of his research:
- In general, the base makeup of DNA differs between species.
- The base composition of DNA extracted from different organs of the same animal is identical.
- The DNA base composition of a specific species does not alter with age, nutritional status, or environmental change.
- In all cellular DNAs, regardless of species, the number of adenosine residues is equal to the number of thymidine residues (that is, A:T) and the number of guanosine residues is equal to the number of cytidine residues (G:C). Therefore, the sum of the purine residues equals the sum of the pyrimidine residues, or A + T = G + C.
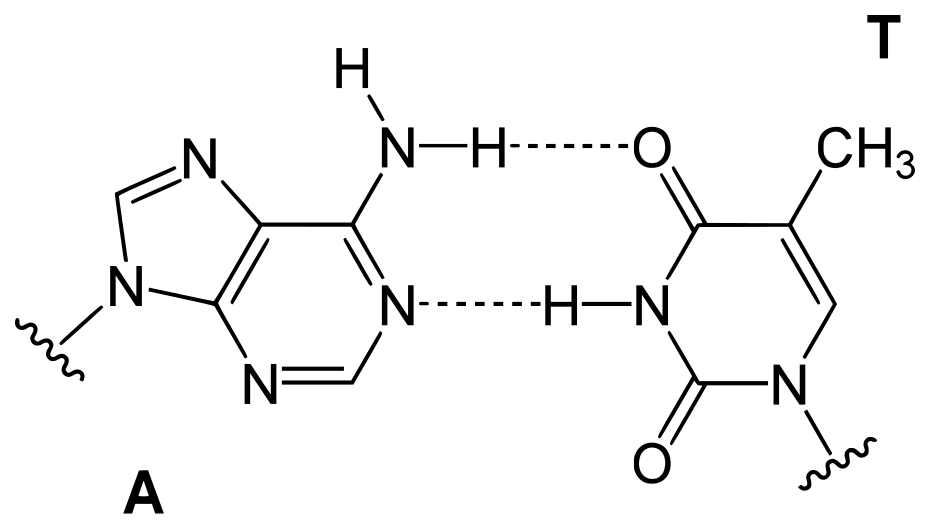
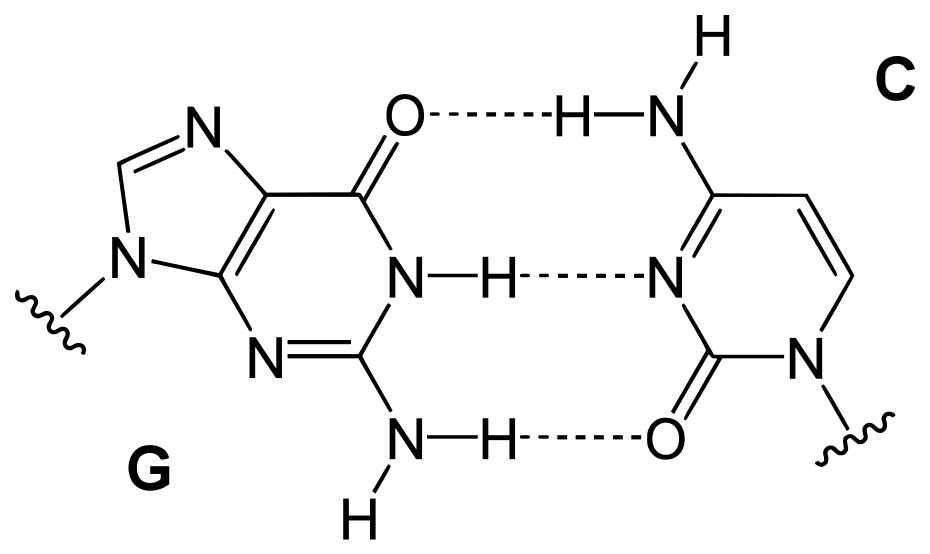
These quantitative links, sometimes known as “Chargaff’s laws,” were validated by a large number of later studies. They were crucial in determining the three-dimensional structure of DNA and provided insight into how genetic information is encoded in DNA and carried from generation to generation.
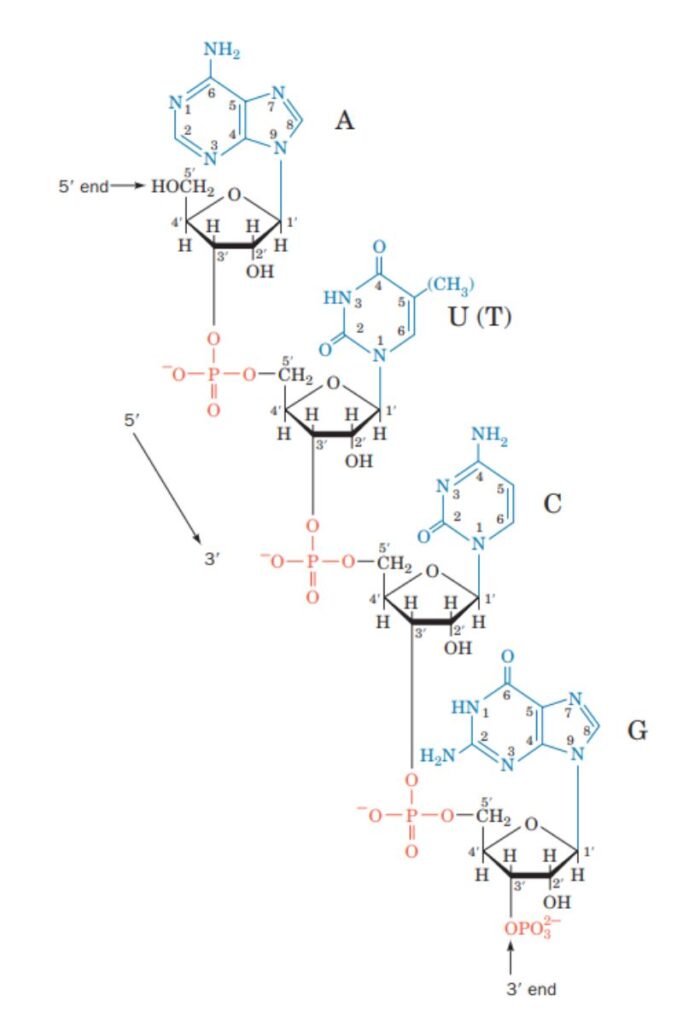
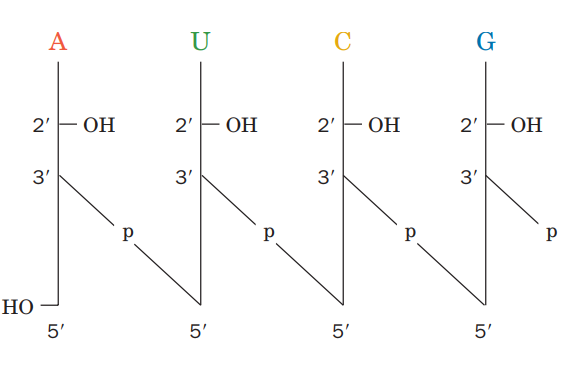
How to write a sequence of DNA?
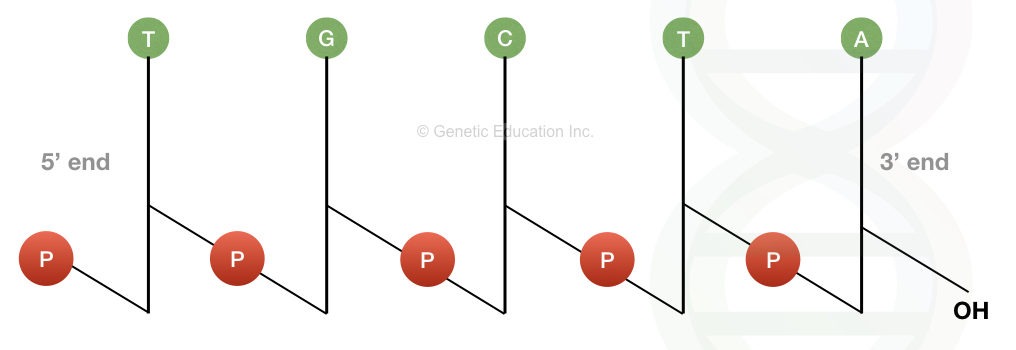
The image above demonstrates that the DNA molecule depicted is a polydeoxyribonucleotide designated as pT-G-C-T-AOH.
Now let us evaluate its performance.
Each deoxyribose is represented by a vertical line. Carbon C1 with a single base is at the top of the line, while carbon C5 is at the bottom. P is the symbol for phosphate, while each nitrogenous base is represented by its own symbol (viz A, T, C and G).
The line crosses through the phosphate at where a 5′ end of one sugar and a 3′ end of another sugar are connected. Based on the above description, the DNA molecule can be written as 5′-pT-G-C-T-AOH or pTpGpCpTpAOH.
In a single-stranded DNA, the left end is always 5′ and the right end is always 3′; consequently, it is written as 5′ to 3′.
If the DNA has six nucleotides, it is referred to as hexadeoxyribonucleotide, etc. In general, however, oligonucleotides are composed of up to 50 nucleotides, whereas polynucleotides have more than 50 nucleotides.
Before we can comprehend the structure of DNA, we must first comprehend the actual model of DNA structure presented by Watson and Crick, and to do so, we must collect DNA evidence.
DNA replication
DNA replication is a fundamental process in which the genetic information stored in DNA is accurately copied to ensure the transmission of genetic material to daughter cells. The replication process involves several key steps and molecular components.
- Unwinding of DNA strands: The double-stranded DNA molecule must first separate, or unwind, to expose the template strands for replication. DNA gyrase, a class II topoisomerase, helps in uncoiling the DNA ahead of the replication fork. Helicase is responsible for further unwinding the DNA at the replication fork.
- Stabilization of unwound DNA: Single-strand binding proteins (SSBs) bind to the single-stranded DNA regions exposed by helicase, preventing re-annealing and maintaining the DNA strands in an unwound state.
- Priming: Primase, an RNA polymerase, synthesizes a short RNA primer complementary to the DNA template. This primer provides the starting point for DNA synthesis.
- DNA synthesis: DNA polymerase takes over after the RNA primer is laid down. DNA polymerase adds nucleotides complementary to the exposed template strands, extending the growing DNA chain in the 5′ to 3′ direction. The synthesis occurs on both strands of the unwound DNA.
- Leading and lagging strands: The DNA synthesis that proceeds continuously in the same direction as the replication fork is called the leading strand. The opposite strand, which requires synthesis in short fragments, is known as the lagging strand. These short fragments are called Okazaki fragments.
- Primer replacement and Okazaki fragment joining: The RNA primers are later replaced with DNA by a special DNA polymerase. Once the RNA primers are removed, DNA ligase joins the adjacent Okazaki fragments on the lagging strand, creating a continuous DNA strand.
- Bidirectional replication: DNA replication occurs bidirectionally from the origin of replication, forming two replication forks that proceed in opposite directions. This process resembles an expanding bubble, with DNA synthesis occurring simultaneously on both strands.
- Proofreading and error correction: DNA polymerase has proofreading activity, allowing it to recognize and correct any mistakes (mutations) it makes during DNA synthesis, thus ensuring high fidelity.
- Semi-conservative nature: DNA replication is semi-conservative, meaning that each newly synthesized DNA molecule consists of one strand from the original DNA molecule (parental strand) and one newly synthesized strand (daughter strand). This was experimentally demonstrated by Meselson and Stahl using heavy (15N) and light (14N) nucleotides, which resulted in DNA of intermediate weight after one round of replication and a mixture of intermediate and light DNA after the second round.
- Timing of replication: DNA replication occurs once during the cell cycle, specifically in the S (synthesis) phase. In meiosis, replication occurs twice, but still only once per original DNA molecule.
Understanding the mechanism of DNA replication is crucial for maintaining genetic integrity and accurate transmission of genetic information from one generation to the next.
Functions of DNA
- Genes and genomes: To suit the little available volume within the cell, genomic DNA is tightly and neatly packed by a process known as DNA condensation. DNA is mostly located in the nucleus of eukaryotic cells, with smaller amounts in mitochondria and chloroplasts. In prokaryotes, DNA is contained within a nucleoid, an irregularly structured substance in the cytoplasm. The genetic information contained within a genome’s genes is referred to as the genotype of an organism. A gene is a unit of heredity and a segment of DNA that controls a certain trait in an organism. The open reading frame and regulatory sequences, like as promoters and enhancers, that control transcription of the open reading frame are components of genes.
- Translation: mRNA synthesises a lengthy chain of amino acids in the cytoplasm by utilising rRNA and tRNA molecules. Proteins are formed by formatting their primary, secondary, tertiary, and quaternary structures.
- Transcription: In the process known as transcription, messenger RNA is created from the replicated DNA. The RNA polymerase synthesises an mRNA molecule from single-stranded DNA and retains all the information necessary for the expression of a gene. Once mRNA is generated, it leaves the nucleus to undergo the translation process.
- Replication: Replication is the process by which DNA molecules are duplicated by enzyme processes. The helicase unwinds the double-stranded DNA, the primase places an RNA primer on the leading strand, and a polymerase synthesises new DNA.
- Extracellular nucleic acids: Bare extracellular DNA (eDNA) is practically ubiquitous in the environment; the majority of it is liberated after cell death. Its concentration in natural aquatic habitats may reach 88 g/L, and its concentration in soil may reach 2 g/L. eDNA may be engaged in horizontal gene transfer, may offer nutrients, and may operate as a buffer to attract or titrate ions or antibiotics. In the biofilms of a number of bacterial species, extracellular DNA functions as an extracellular matrix component. It may function as a recognition factor to govern the attachment and dispersal of specific cell types in the biofilm, contribute to biofilm development, and contribute to the biofilm’s physical robustness and resistance to biological stress.
- Neutrophil extracellular traps: Neutrophil extracellular traps (NETs) are networks of extracellular fibres consisting mostly of DNA that enable neutrophils, a kind of white blood cell, to destroy extracellular pathogens while limiting damage to the host cells.
Use of DNA in different fields
Genetic engineering
- As an example, phenol-chloroform extraction can be used to isolate DNA from organisms, and restriction digests and the polymerase chain reaction can be used to alter it in the laboratory.
- These techniques in recombinant DNA technology are heavily utilised by modern biology and biochemistry.
- Recombinant DNA is a sequence of DNA that has been synthesised by combining different DNA sequences. Using a viral vector, plasmids or the suitable format can be turned into organisms.
- Produced genetically modified organisms can be used to make recombinant proteins, in medical research, or for agricultural purposes.
DNA profiling
- Using the DNA in blood, sperm, skin, saliva, or hair collected at a crime scene, forensic scientists can identify a culprit with matching DNA.
- This procedure is properly known as DNA fingerprinting or DNA profiling.
- In DNA profiling, the lengths of variable regions of repetitive DNA, such as minisatellites and short tandem repeats, are compared between individuals.
- Typically, this methodology is quite reliable for identifying DNA matches.
DNA enzymes or catalytic DNA
- Deoxyribozymes, commonly known as DNAzymes or catalytic DNA, were identified for the first time in 1994.
- They are predominantly single-stranded DNA sequences extracted from a huge pool of random DNA sequences by in vitro selection or systematic evolution of ligands by exponential enrichment (SELEX).
- DNAzymes catalyse several chemical reactions, such as RNA-DNA cleavage, RNA-DNA ligation, amino acid phosphorylation-dephosphorylation, carbon-carbon bond creation, etc.
- DNAzymes can increase the catalytic rate of chemical reactions by a factor of up to 100,000,000,000 above uncatalyzed reactions.
Bioinformatics
- Bioinformatics is the development of techniques for storing, mining, searching, and manipulating biological data, such as DNA nucleic acid sequence data.
- In particular, string searching algorithms, machine learning, and database theory have led to widespread advancements in computer science.
- To search for specific sequences of nucleotides, string searching or matching algorithms, which locate an occurrence of a sequence of letters inside a larger sequence of letters, were developed.
- The DNA sequence can be matched with other DNA sequences in order to discover homologous sequences and locate the mutations that distinguish them.
- The study of evolutionary relationships and protein function employs these approaches, particularly multiple sequence alignment.
DNA nanotechnology
- DNA nanotechnology employs the distinctive molecular recognition properties of DNA and other nucleic acids to produce self-assembling branching DNA complexes with advantageous qualities.
- Thus, DNA is utilised as a construction material rather than a biological information carrier.
- This has led to the fabrication of two-dimensional periodic lattices (both tile-based and utilising the DNA origami approach) and three-dimensional polyhedral structures.
- Additionally, nanomechanical devices and algorithmic self-assembly have been demonstrated, and these DNA structures have been utilised as a template for the arrangement of other molecules, including gold nanoparticles and streptavidin proteins.
History and anthropology
- Because DNA accumulates mutations over time, which are then inherited, it carries historical information, and geneticists can infer the evolutionary history of species, their phylogeny, by comparing DNA sequences.
- In evolutionary biology, phylogenetics is a potent tool.
- By comparing DNA sequences within a species, population geneticists can determine the history of specific populations. This can be utilised in anthropology and ecological genetics research.
Information storage
- DNA has immense potential as a data storage medium since it has a far higher storage density than electronic devices.
- However, expensive costs, slow read and write times (memory latency), and inadequate dependability have impeded its practical application.
Different types of DNA based on their topology
Circular DNA, linear DNA, and supercoiled DNA are the three kinds of DNA found in nature, depending on their topology.
Circular DNA
- Both ends of the double-stranded DNA are linked to form a closed loop in circular DNA.
- The circular DNA is devoid of supercoiling (without any twists or writes).
- In prokaryotes, circular DNA is typically present as a plasmid.
- Additionally, some eukaryotic organelles, including mitochondria and chloroplast, contain circular DNA.
- When the virus enters the nucleus of the host cell, it is transformed into covalently closed circular DNA, a unique form of circular DNA (cccDNA).
Different types of DNA based on their location
On the basis of where the DNA is located, all organisms include two basic forms of DNA: nuclear DNA and cytoplasmic DNA.
Nuclear DNA
- In all eukaryotes, the bulk of DNA is located in the nucleus of the cell. DNA in the nucleus contains all essential genes. Metaphase chromosomes contain the entirety of the nuclear DNA.
- During cell division, it is divided and passed on to progeny.
Cytoplasmic DNA
- DNA is also found in the cytoplasm of the cell, but only in organelles enclosed by membranes.
- Mitochondria and chloroplasts carry their own DNA, abbreviated as mtDNA and cpDNA, respectively.
- Both possess specialised replication and gene expression machinery.
- Mitochondria and chloroplasts are considered to have been free-living prokaryotes in the past.
- Furthermore, cytoplasmic DNA contains critical genes; mutations in these genes result in fatal illnesses.
Linear DNA
- The ends of linear DNA are unbound. A portion of the short and free DNA fragments found in the cell are linear DNA.
- Some prokaryotes possess linear DNA as well.
Supercoiled DNA
- When the long chain of DNA twists around itself, a complicated form of DNA known as supercoiled DNA is formed.
- In eukaryotic genomes, the supercoiled form of DNA is more widespread, allowing the lengthy DNA to fit inside the cell nucleus.
- Additionally, supercoiling helps DNA arrange itself on chromosomes by interacting with certain proteins.
How DNA Is Arranged in the Cell?
DNA is a functional molecule; it must be replicated when a cell is ready to divide and “read” to produce the molecules, such as proteins, that carry out the cell’s functions. Because of this, the DNA is packaged and protected in very specific ways. Additionally, DNA molecules can be quite lengthy. The DNA molecules in a single human cell would reach a length of approximately 2 meters if stretched end-to-end. To fit and function within a structure (the cell) that is not visible to the unaided eye, the DNA of a cell must be packaged in a highly organized fashion. In many ways, the chromosomes of prokaryotes are much simpler than those of eukaryotes. Most prokaryotes contain a single circular chromosome, which is located in the nucleoid of the cytoplasm.
Escherichia coli, one of the most extensively studied prokaryotes, has a genome of 4.6 million base pairs, which, if extended, would measure approximately 1.6 mm. How then does this fit within a bacterial cell? In a process known as supercoiling, the DNA is coiled beyond the double helix. Some proteins are known to play a role in supercoiling, while other proteins and enzymes contribute to the maintenance of the supercoiled structure.
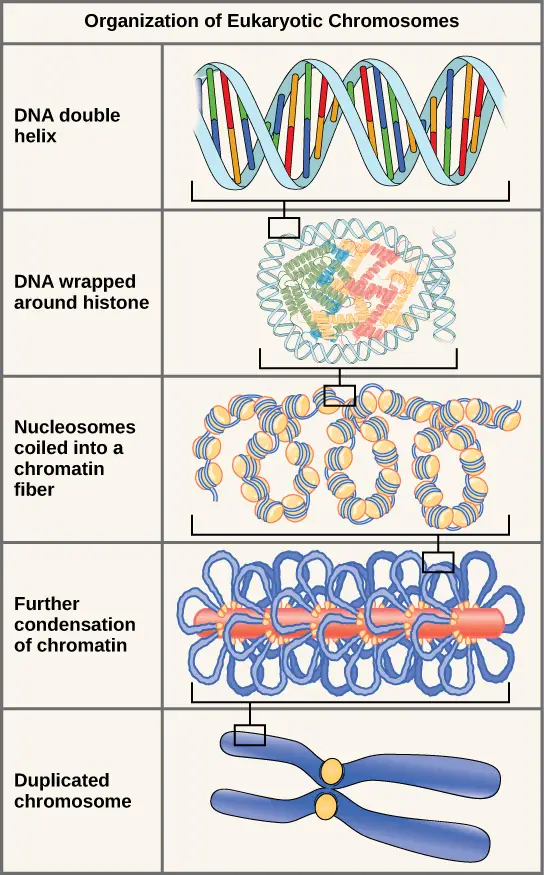
Eukaryotes, whose chromosomes each consist of a linear DNA molecule, employ a distinct packaging strategy to accommodate their DNA within the nucleus. At the most fundamental level, DNA is coiled around proteins called histones to form nucleosomes. DNA is firmly wrapped around the histone core. This nucleosome is connected to the next one by a brief, histone-free DNA strand. This structure is also known as the “beads on a string” structure; the nucleosomes are the “beads” and the DNA segments between them are the “string.” The DNA-wrapped nucleosomes form a 30-nm-wide fiber by stacking tightly to form a 30-nm-wide fiber. This fiber is further wound into a structure that is thicker and more compact. At the metaphase stage of mitosis, when the chromosomes are aligned in the middle of the cell, they are at their densest. They have a width of approximately 700 nm and are associated with scaffold proteins.
In interphase, the phase of the cell cycle between mitoses during which the chromosomes are decondensed, eukaryotic chromosomes have two distinct regions that can be distinguished through staining. There is a densely packed, darkly stained region and a less dense region. The regions that stain darkly typically contain inactive genes and are located in the centromere and telomere regions. The faintly stained regions typically contain active genes, with DNA packaged around nucleosomes but not compacted further.
Key Terms and Points
- Nucleotide: It is the basic building block of DNA and consists of a base (adenine, guanine, thymine, or cytosine), a sugar, and a phosphate group.
- Nucleoside: It is formed when a base is attached to a sugar, such as adenosine, guanosine, thymidine, or cytidine.
- Sugar-phosphate backbone: The backbone of DNA is made up of alternating sugar (deoxyribose) and phosphate molecules. Phosphodiester bonds link the 5′ phosphate of one nucleotide to the 3′ hydroxyl group of the next nucleotide.
- Base: DNA bases can be purines (adenine and guanine) or pyrimidines (thymine and cytosine). Purines are larger bases with two rings, while pyrimidines are smaller with one ring.
- Uracil: In RNA, thymine is replaced by uracil as one of the bases.
- Acidity of DNA: The phosphate group in the nucleotide gives DNA its acidic nature.
- Double helix: DNA exists as a double-stranded molecule with a helical structure. This is known as the Watson-Crick model of DNA structure.
- Antiparallel: The two strands of DNA run in opposite directions. One strand goes from 5′ to 3′, while the other goes from 3′ to 5′.
- Helix: The double-stranded DNA is coiled into a spiral shape.
- Base pairing specificity: Adenine (A) forms two hydrogen bonds with thymine (T), while guanine (G) forms three hydrogen bonds with cytosine (C). A always pairs with T, and G always pairs with C.
- GC content: DNA molecules with a high guanine-cytosine (GC) content have a higher number of hydrogen bonds and are more stable and harder to denature.
- Complementary strands: The two strands of DNA are complementary to each other, with base pairing between A-T and G-C.
- Denaturation: It is the separation of the two complementary DNA strands, often caused by high temperature.
- Reannealing/Hybridization/Annealing: It refers to the process of the complementary DNA strands coming back together. Reannealing occurs after denaturation, while hybridization or annealing is the process of complementary strands joining together. This is used in techniques like PCR (polymerase chain reaction), Southern blotting, and plasmid ligation.
FAQ
How do DNA strands are Joint Together?
Adenine and thymine are linked by two hydrogen bonds, but cytosine and guanine are linked by three hydrogen bonds. The DNA molecule is stable due to the stacking of the bases and the hydrophobic interaction between the bases.
What is Complementary Base-Pairing?
To assist the bases in each base pair fit into the double helix, the two DNA strands of the double helix run in opposite directions (antiparallel to each other). This indicates that the nucleotides in each DNA strand are perfectly complementary to those in the other strand.
The complementary base-pairing of A with T and G with C optimises double-stranded DNA’s energy levels. In this double helix configuration, the width of each base pair remains constant, meaning that the distance between the sugar-phosphate backbones is maintained along the entire length of the DNA molecule. Twists or turns in the two sugar-phosphate backbones of the double helix occur every ten base pairs, maximising the packing efficiency.
The DNA double helix’s two polynucleotide strands provide a straightforward basis for copying the molecule’s information. Upon separation, each of the two strands serves as a template for duplicating the DNA molecule exactly or identically.
Who Discovered the Structure of DNA?
The structure of DNA was discovered by James Watson and Francis Crick, along with significant contributions from Rosalind Franklin and Maurice Wilkins. In 1953, Watson and Crick proposed the double helix structure of DNA, which is now widely accepted. Their discovery was based on X-ray crystallography images of DNA taken by Franklin and Wilkins, which provided crucial insights into its structure. Watson and Crick’s model explained how DNA’s two strands are connected by base pairs (A with T and C with G) and held together by hydrogen bonds. This groundbreaking discovery revolutionized our understanding of genetics and laid the foundation for modern molecular biology.
What is DNA?
DNA stands for Deoxyribonucleic Acid. It is a molecule found in cells that contains the genetic instructions for the development, functioning, and reproduction of all living organisms.
What is the structure of DNA?
DNA has a double-helix structure, resembling a twisted ladder. It consists of two strands made up of nucleotides, which are chemical building blocks. The nucleotides are composed of a sugar molecule, a phosphate group, and one of four nitrogenous bases: adenine (A), thymine (T), cytosine (C), and guanine (G).
What is the function of DNA?
The primary function of DNA is to store and transmit genetic information. It serves as a blueprint for the synthesis of proteins, which are essential for the structure and function of cells and organisms.
Where is DNA found?
DNA is found in the nucleus of eukaryotic cells, which include plants, animals, fungi, and protists. It is also found in the cytoplasm of prokaryotic cells, such as bacteria.
How is DNA replicated?
DNA replication is the process by which DNA makes an identical copy of itself. It occurs during cell division and involves the separation of the DNA strands, followed by the synthesis of two new complementary strands using the existing strands as templates.
What is the role of DNA in inheritance?
DNA carries the genetic information that is passed from parents to offspring during reproduction. It contains genes, which are specific segments of DNA that code for particular traits or characteristics.
Can DNA be modified or altered?
Yes, DNA can be modified or altered through processes such as mutations. Mutations are changes in the DNA sequence that can occur spontaneously or due to environmental factors. They can have various effects on an organism, ranging from no noticeable change to significant impacts on traits or health.
How is DNA used in forensic science?
DNA profiling, also known as DNA fingerprinting, is used in forensic science to identify individuals and establish biological relationships. DNA samples collected from crime scenes or other sources can be compared to known DNA samples to determine matches or exclusions.
What is the Human Genome Project?
The Human Genome Project was an international scientific research project that aimed to map and sequence the entire human genome. It was completed in 2003 and provided a blueprint of the genetic information encoded in human DNA, leading to advancements in medicine, genetics, and biotechnology.
How is DNA used in genetic engineering?
Genetic engineering involves manipulating the DNA of organisms to introduce or modify specific traits. DNA can be extracted, modified, and reinserted into an organism to change its characteristics or create new traits. This technology has applications in fields such as agriculture, medicine, and biotechnology.
References
- Alberts B, Johnson A, Lewis J, et al. Molecular Biology of the Cell. 4th edition. New York: Garland Science; 2002. The Structure and Function of DNA. Available from: https://www.ncbi.nlm.nih.gov/books/NBK26821/
- https://profiles.nlm.nih.gov/spotlight/sc/feature/doublehelix
- https://www.mun.ca/biology/scarr/Watson-Crick_Model.html
- https://www.nature.com/scitable/topicpage/discovery-of-dna-structure-and-function-watson-397/
- https://www.khanacademy.org/test-prep/mcat/biomolecules/dna/a/dna-structure-and-function
- https://www.khanacademy.org/science/ap-biology/gene-expression-and-regulation/replication/a/hs-dna-structure-and-replication-review
- https://www.medicalnewstoday.com/articles/319818
- https://www.clinicalkey.com/#!/content/book/3-s2.0-B9781437706963000029
- https://www.osmosis.org/learn/DNA_structure
- https://febs.onlinelibrary.wiley.com/doi/10.1111/febs.13307
- https://ib.bioninja.com.au/standard-level/topic-2-molecular-biology/26-structure-of-dna-and-rna/dna-structure.html
- https://biologydictionary.net/dna-structure/
- https://www.ufrgs.br/imunovet/molecular_immunology/DNAstructureanalysis.html
- https://www.ndsu.edu/pubweb/~mcclean/plsc731/dna/dna4.htm
- https://www.news-medical.net/life-sciences/Structure-of-DNA.aspx
- https://www.compoundchem.com/2015/03/24/dna/
- https://www.genome.gov/genetics-glossary/Deoxyribonucleic-Acid#:~:text=DNA%20is%20made%20of%20two,)%20or%20thymine%20(T).
- https://microbenotes.com/dna-structure-properties-types-and-functions/
- https://www.umass.edu/molvis/tutorials/dna/
- https://medlineplus.gov/genetics/understanding/basics/dna/
- https://www.visiblebody.com/learn/biology/dna-chromosomes/dna-structure
- https://en.wikipedia.org/wiki/DNA
- https://www.chemguide.co.uk/organicprops/aminoacids/dna1.html
- https://www.livescience.com/37247-dna.html
