Table of Contents
- DNA methylation is a process where a methyl group is added to a cytosine residue in DNA.
- Methyl groups are added by enzymes called DNA methyltransferases (DNMTs).
- DNA methylation can occur at specific sites called CpG dinucleotides, where a cytosine is followed by a guanine.
- DNA methylation can regulate gene expression by affecting the binding of transcription factors to DNA.
- There are two types of DNA methylation: de novo methylation, which adds methyl groups to previously unmethylated DNA, and maintenance methylation, which preserves existing methylation patterns during DNA replication.
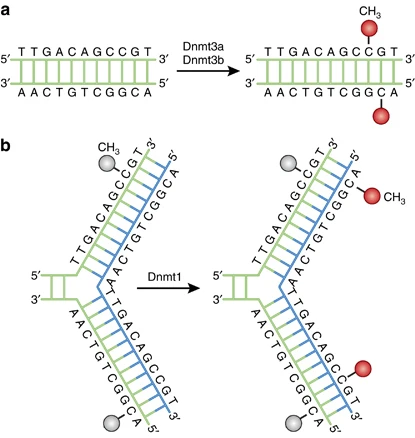
- De novo DNA methylation is carried out by DNMT3A and DNMT3B enzymes.
- Maintenance DNA methylation is performed by DNMT1, which copies the existing methylation pattern onto the newly synthesized DNA strand during replication.
- DNA demethylation is the removal of methyl groups from DNA. It can occur through passive demethylation during cell division or active demethylation involving enzymatic reactions.
- Various proteins called methyl-binding proteins (MBPs), including MBD proteins, UHRF proteins, and zinc-finger proteins, can recognize and bind to methylated DNA.
- Methyl-binding proteins can influence gene expression by either repressing transcription or inhibiting the binding of transcriptional activators.
- DNA methylation patterns can be heritable and play important roles in development, genomic imprinting, and X-chromosome inactivation.
- Dysregulation of DNA methylation is associated with various diseases, including cancer and neurological disorders.
What is DNA Methylation?
- DNA methylation is the biological addition of methyl groups to the DNA molecule. Methylation can alter a DNA segment’s activity without altering its sequence.
- DNA methylation, when present in a gene promoter, typically inhibits gene transcription. DNA methylation is required for normal development in mammals and is associated with a number of important processes, such as genomic imprinting, X-chromosome inactivation, repression of transposable elements, ageing, and carcinogenesis.
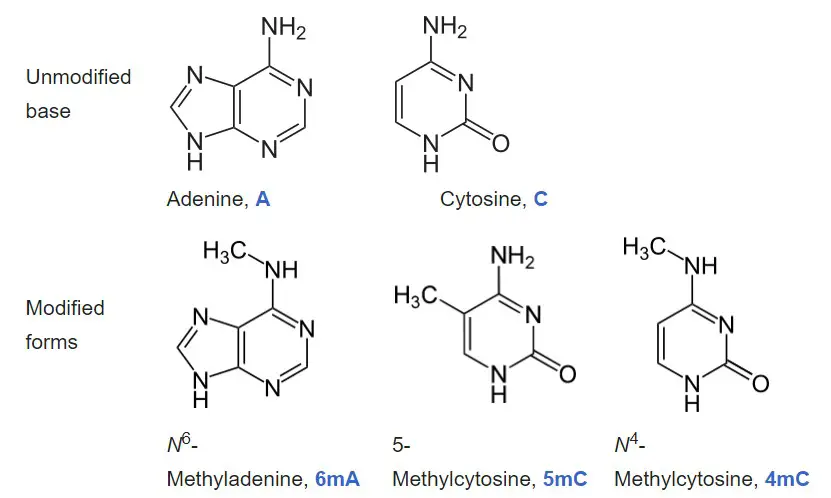
- Adenine and cytosine are the two nucleobases that undergo natural, enzymatic DNA methylation as of 2016. N6-methyladenine, 5-methylcytosine, and N4-methylcytosine are the modified bases.
- Cytosine and adenine are two of the four bases of DNA that can be methylated. Cytosine methylation is prevalent in both eukaryotes and prokaryotes, despite the fact that the rate of cytosine DNA methylation can vary substantially between species: 14% of cytosines are methylated in Arabidopsis thaliana, 4% to 8% in Physarum, 7.6% in Mus musculus, 2.3% in Escherichia coli, 0.03% in Drosophila, 0.006% in Dictyostelium and virtually none (0.0002 to 0.0003%) in Caenorhabditis or fungi such as Saccharomyces cerevisiae and S. pombe (but not N. crassa).
- Adenine methylation has been observed in bacterial, plant, and, more recently, mammalian DNA, but has received much less attention.
- Methylation of cytosine to produce 5-methylcytosine occurs at the same 5 position on the pyrimidine ring where the methyl group of the DNA base thymine is located; this distinguishes thymine from the analogous RNA base uracil, which does not contain a methyl group. 5-methylcytosine is converted to thymine by spontaneous deamination.
- This results in an improper T:G match. Repair mechanisms then restore it to its original C:G pair; alternatively, they may substitute A for G, transforming the original C:G pair into a T:A pair, thereby altering a base and introducing a mutation.
- As thymine is a DNA base, this misincorporated base will not be corrected during DNA replication. If the mismatch is not repaired and the cell enters the cell cycle, the T-containing strand will be complemented by an A in one of the progeny cells, resulting in a permanent mutation.
- The nearly exclusive use of thymine in DNA and uracil in RNA may have evolved as an error-control mechanism to facilitate the elimination of uracils generated by the spontaneous deamination of cytosine.
- Several lines of evidence suggest that DNA methylation and many of its contemporary DNA methyltransferases evolved from primitive RNA methylation activity in the early world.
- DNA methylation occurs in three distinct sequence contexts in plants and other organisms: CG (or CpG), CHG, and CHH (where H corresponds to A, T, or C).
- DNA methylation occurs almost exclusively in CpG dinucleotides in mammals, with cytosines on both strands typically being methylated. However, non-CpG methylation can be observed in embryonic stem cells and has also been linked to neural development. In addition, non-CpG methylation has been observed in hematopoietic progenitor cells, primarily in the context of CpApC sequences.
DNA Methylation Definition
DNA methylation is an epigenetic process in which a methyl group (CH3) is added to the DNA molecule. This modification primarily occurs at the 5-carbon position of the cytosine ring, resulting in the formation of 5-methylcytosine (5-mC). It is considered one of the major mechanisms for regulating gene expression and can impact the function of genes by influencing their transcriptional activity. DNA methylation plays a crucial role in various biological processes, including development, differentiation, genomic imprinting, and X-chromosome inactivation.
or
DNA methylation is a fundamental biological process that involves the addition of methyl groups to the DNA molecule. This modification occurs without altering the underlying DNA sequence. When present in gene promoters, DNA methylation usually functions to suppress gene transcription. In mammals, DNA methylation plays a crucial role in various vital processes, such as genomic imprinting, X-chromosome inactivation, repression of transposable elements, aging, and the development of cancer. It is an essential mechanism for normal development and the regulation of gene activity.
Principle of DNA Methylation
Although almost all cells in an organism possess the same genetic material, not all genes are expressed by all cell types at the same time. This variation in gene expression is regulated by epigenetic mechanisms, which are critical in regulating gene expression profiles across different cells and organs in multicellular organisms. DNA methylation, which includes the direct chemical change of DNA, is one such key epigenetic mechanism.
During development, DNA methylation patterns in the genome vary dynamically, comprising both de novo DNA methylation and demethylation mechanisms. These changes aid in the formation of distinct and persistent DNA methylation patterns in differentiated cells, thereby regulating tissue-specific gene transcription.
Further information:
- The addition of a methyl group to the DNA molecule, most notably at the 5-carbon position of cytosine residues, results in 5-methylcytosine (5-mC).
- DNA methyltransferases (DNMTs) are a family of enzymes that methylate DNA.
- De novo DNA methylation is carried out by DNMT3a and DNMT3b, which establishes methylation patterns during development.
- DNMT1 is important in the maintenance of pre-existing DNA methylation patterns during DNA replication.
- DNA demethylation, or the removal of methyl groups from DNA, can happen passively or actively.
- When newly produced strands lack methyl groups, passive DNA demethylation occurs during replication.
- Enzymatic mechanisms, such as TET-mediated oxidation, are used in active DNA demethylation to remove methyl groups.
- Environmental variables, aging, and disease can all have an impact on DNA methylation patterns.
- Several disorders, including cancer, have been linked to abnormal DNA methylation patterns, such as global hypomethylation or gene-specific hypermethylation.
- DNA methylation can influence gene expression by preventing transcription factors from binding or recruiting proteins involved in gene silence.
- DNA methylation is important in genomic imprinting, which is the expression of certain genes based on their parental origin.
- During development, changes in DNA methylation patterns are required for cellular differentiation and the creation of specialized cell types.
Location Of Dna Methylation
- DNA methylation is more common on cytosines that come before guanine nucleotides, or CpG sites.
- Although the brain contains significant levels of DNA methylation, 5mC (methylated cytosine) accounts for less than 1% of nucleic acids in the human genome.
- Except for CpG islands, the majority of DNA methylation occurs at CpG sites.
- CpG islands are DNA sequences with a high CpG density that are commonly found in gene promoters. They are not commonly methylated.
- CpG islands are linked to gene expression regulation by enhancing chromatin accessibility and transcription factor binding.
- CpG island methylation results in persistent gene silencing, which is critical for imprinting and controlling gene expression during development and differentiation.
- CpG islands associated with transcription start sites seldom exhibit tissue-specific methylation patterns, whereas CpG island coasts (regions up to 2 kb distant) do.
- To suppress potentially damaging genetic elements, intergenic regions, which contain transposable and viral elements, are extensively methylated.
- Methylation of the gene body (regions past the first exon) is linked to increased gene expression in dividing cells, but its effect in nondividing cells, such as the brain, is unknown.
- Non-CpG methylation has been seen in mouse and human embryonic stem cells, but it disappears in adult tissues. Its function is yet not completely known.
Intergenic Regions
- Around 45% of the mammalian genome is made up of transposable and viral elements that are silenced by DNA methylation.
- Methylation restricts their expression since these elements can be damaging by disrupting genes and generating DNA alterations. Throughout life, the intracisternal A particle (IAP), a retrovirus in mice, is extensively methylated, including during gametogenesis, development, and adulthood.
- Even during embryonic stages, when the remainder of the genome is relatively hypomethylated, the enzyme Dnmt1 maintains IAP suppression.
- IAP elements can be expressed when Dnmt1 is reduced, resulting in widespread hypomethylation. As a result, DNA methylation is important in suppressing the expression of potentially dangerous genetic sequences within intergenic regions.
CpG Islands
- CpG islands are DNA regions with a high CpG density that are typically unmethylated.
- The majority of gene promoters, particularly those for housekeeping genes, are found on CpG islands.
- CpG islands have evolved to play an important role in gene expression by controlling chromatin shape and transcription factor binding.
- CpG islands have fewer nucleosomes than other DNA locations and frequently have histone modifications that boost gene expression.
- CpG islands make DNA more accessible and encourage transcription factor binding.
- Methylation of CpG islands results in long-term suppression of gene expression.
- During gametogenesis and early embryonic development, CpG islands undergo differential methylation, which plays an important role in imprinting.
- CpG island DNA methylation influences gene expression throughout development and differentiation.
- Tissue-specific methylation patterns on CpG islands are rare, but tissue-specific methylation patterns on CpG island coastlines are highly conserved.
- CpG shore methylation is linked to decreased gene expression.
- CpG islands’ role in gene expression regulation is yet unknown, and further research is needed to discover the degree of their impact.
Gene Body
- CpG sites within the mammalian genome are typically methylated, including genes themselves.
- Methylation of the first exon or promoter region leads to gene silencing.
- Gene body refers to the region of the gene beyond the first exon.
- Methylation of the gene body is associated with higher gene expression in dividing cells.
- In slowly dividing and nondividing cells like the brain, gene body methylation does not increase gene expression.
- Methylation of non-CpG sites within gene bodies in the murine frontal cortex is negatively correlated with gene expression.
- The role of DNA methylation in the gene body in gene regulation is still not well understood.
Types of DNA methylation
There are several types of DNA methylation that occur in different contexts and have distinct functions. The main types of DNA methylation include:
- CpG Methylation: CpG methylation refers to the addition of a methyl group to the carbon atom in the cytosine residue of a CpG dinucleotide. CpG sites are regions in the DNA where a cytosine is followed by a guanine nucleotide. CpG methylation is the most well-studied and common form of DNA methylation in mammals.
- Non-CpG Methylation: Non-CpG methylation involves the methylation of cytosines in sequence contexts other than CpG, such as CpA, CpT, and CpC. Non-CpG methylation is relatively rare in most tissues, but it has been observed in certain cell types, including embryonic stem cells and neurons.
- Promoter Methylation: Promoter methylation occurs at the promoter region of a gene, which is crucial for regulating gene expression. Methylation of CpG sites in gene promoters often leads to gene silencing by inhibiting the binding of transcription factors and other regulatory proteins.
- Gene Body Methylation: Gene body methylation refers to the methylation that occurs within the transcribed region of a gene, including the coding sequence. Unlike promoter methylation, gene body methylation is generally associated with active gene expression, and its precise role is still being investigated.
- Imprinting Methylation: Imprinting methylation is a unique form of DNA methylation that occurs at specific loci and is involved in parent-specific gene expression. Imprinted genes are marked with methylation on one parental allele, leading to the silencing or activation of specific genes depending on their parental origin.
- Global Methylation: Global methylation refers to the overall level of DNA methylation across the entire genome. It represents the average extent of methylation throughout the genome and is often measured as a global indicator of DNA methylation status.
These different types of DNA methylation contribute to the regulation of gene expression, cellular differentiation, development, and other biological processes. Understanding their roles and dynamics is crucial for unraveling the complex mechanisms of epigenetic regulation.
Mechanism of DNA Methylation
DNA methylation involves three classes of enzymes: writers, erasers, and readers, which play distinct roles in establishing, removing, and interpreting DNA methylation marks.
- Writers: These enzymes, known as DNA methyltransferases (DNMTs), are responsible for adding methyl groups to cytosine residues. They catalyze the transfer of a methyl group onto the carbon atom of the cytosine base, resulting in 5-methylcytosine (5mC). DNMTs are involved in establishing and maintaining DNA methylation patterns.
- Erasers: These enzymes are responsible for modifying or removing the methyl group from 5mC, leading to demethylation of DNA. One well-known eraser enzyme is the ten-eleven translocation (TET) family of proteins, which oxidize 5mC to initiate the demethylation process. Erasers are involved in dynamic regulation of DNA methylation patterns.
- Readers: These proteins recognize and bind to methylated DNA, influencing gene expression and chromatin structure. They can either promote gene repression or activation depending on the specific reader protein and context. Readers include proteins with specialized domains such as methyl-CpG-binding domain (MBD) proteins or zinc finger proteins. They interpret the presence of methyl groups and recruit other proteins to modulate gene expression.
By working together, these three classes of enzymes control the establishment, removal, and interpretation of DNA methylation marks, ultimately playing a crucial role in gene regulation and cellular processes.
1. Writing DNA Methylation: the Dnmts
- Three members of the Dnmt family are Dnmt1, Dnmt3a, and Dnmt3b, which add methyl groups to DNA.
- Dnmt1 is highly expressed in mammalian tissues, including the brain. It preferentially methylates hemimethylated DNA and is involved in DNA replication and maintenance of methylation patterns.
- Knockout of Dnmt1 in mice leads to embryonic lethality and loss of DNA methylation.
- Dnmt3a and Dnmt3b are de novo Dnmts that can methylate both native and synthetic DNA. Dnmt3b is poorly expressed in most tissues, while Dnmt3a is expressed ubiquitously.
- Knockout of Dnmt3b is embryonic lethal, while Dnmt3a knockout mice survive for a short period after birth.
- Dnmt3L is a non-catalytic protein that associates with Dnmt3a and Dnmt3b, stimulating their methyltransferase activity. It is mainly expressed in early development and germ cells.
- Dnmt3L is involved in genomic imprinting, retrotransposon methylation, and X chromosome compaction. It is downregulated during neuronal differentiation and not present in the postnatal brain.
Writing DNA Methylation: Targeting De Novo DNA Methylation
- The mechanisms by which de novo Dnmts target specific genomic areas remain unknown.
- Dnmt3a and Dnmt3b have the ability to bind to DNA via a conserved PWWP domain.
- One theory proposes that RNA interference (RNAi) mechanisms target Dnmts to silence specific DNA regions, however evidence in mammalian cells is lacking.
- Transcription factors may influence de novo DNA methylation by recruiting Dnmts for methylation or guarding against DNA methylation.
- To localize methylation to DNA, Dnmts can bind to transcription factors or components of repressor complexes.
- Transcription factor binding, particularly in CpG islands, can help protect CpG sites from de novo methylation.
- Mutations in transcription factor binding sites can cause CpG islands to lose their unmethylated status.
- Downregulation of transcription factors caused by differentiation can expose CpG sites, making them vulnerable to DNA methylation.
- These pathways are most likely working in tandem to create de novo DNA methylation.
- Specific transcription factors may recruit Dnmt3a and Dnmt3b to promoters, or they may methylate CpG sites throughout the genome that are not protected by transcription factor binding.
2. Erasing DNA Methylation
- DNA demethylation can be passive or active.
- Passive DNA demethylation occurs in dividing cells when newly incorporated cytosine remains unmethylated due to inhibition or dysfunction of Dnmt1.
- Active DNA demethylation can occur in both dividing and nondividing cells and requires enzymatic reactions to process 5mC and revert it back to naked cytosine.
- There is no known mechanism in mammalian cells to directly cleave the carbon-to-carbon bond connecting cytosine and the methyl group.
- Active DNA demethylation involves chemical modifications of 5mC, including deamination and oxidation reactions, to generate intermediates recognized by the base excision repair (BER) pathway.
- The BER pathway replaces the modified bases with naked cytosine.
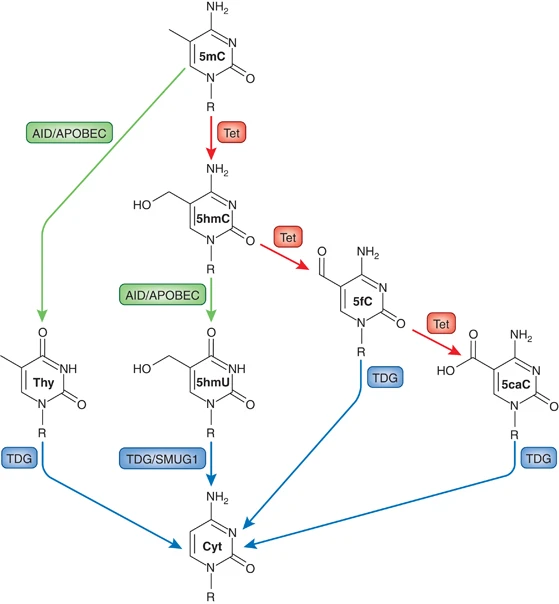
- Several mechanisms of active DNA demethylation have been proposed, including deamination of 5mC by AID/APOBEC enzymes and hydroxylation of 5mC by Tet enzymes.
- AID/APOBEC-mediated deamination converts 5mC to thymine, inducing the BER pathway.
- Tet enzymes add a hydroxyl group to 5mC, forming 5hmC, which can be further oxidized or deaminated.
- Thymine DNA glycosylase (TDG) is essential for the BER pathway and cleaves off the modified residues during DNA demethylation.
- TDG and SMUG1, both BER enzymes, are involved in DNA demethylation.
- The complexity of active DNA demethylation pathways and enzymes contributes to scientific debate in the field.
3. Reading DNA Methylation
- DNA methylation can reduce gene expression by impairing the binding of transcriptional activators.
- DNA methylation is recognized by three families of proteins: MBD proteins, UHRF proteins, and zinc-finger proteins.
- MBD proteins have a conserved methyl-CpG-binding domain (MBD) that gives them a higher affinity for single methylated CpG sites.
- The MBD family includes MeCP2, MBD1, MBD2, MBD3, and MBD4. They can directly bind to methylated DNA and contain a transcriptional repression domain (TRD) that allows them to bind to repressor complexes.
- MeCP2 also plays a role in maintaining DNA methylation by binding to Dnmt1 and recruiting it to hemimethylated DNA for maintenance methylation.
- UHRF proteins, including UHRF1 and UHRF2, bind to methylated cytosines via a SET- and RING-associated DNA-binding domain.
- UHRF proteins primarily function by binding to Dnmt1 and targeting it to hemimethylated DNA, ensuring DNA methylation is maintained during DNA replication.
- UHRF1 interacts closely with Dnmt1, and its deletion leads to embryonic lethality.
- Zinc-finger proteins, including Kaiso, ZBTB4, and ZBTB38, bind to methylated DNA using a zinc-finger domain.
- Kaiso and ZBTB4 have preferential binding for a sequence motif lacking methylcytosine, but they can also recognize methylcytosine.
- Zinc-finger domain proteins repress transcription in a DNA methylation-dependent manner.
- MBD proteins, UHRF proteins, and zinc-finger proteins all contribute to the recognition of DNA methylation and play a role in gene expression regulation.
Summarized Steps of DNA Methylation
DNA methylation is a fundamental epigenetic mechanism that involves the addition of a methyl group to the DNA molecule. The basic mechanism of DNA methylation can be summarized as follows:
- DNA methylation primarily occurs at cytosine residues in the DNA sequence, specifically at CpG sites, where a cytosine nucleotide is followed by a guanine nucleotide.
- The process of DNA methylation is catalyzed by enzymes called DNA methyltransferases (DNMTs). The most well-known DNMT is DNMT1, which is responsible for maintaining DNA methylation patterns during cell division.
- DNMTs add a methyl group (-CH3) to the carbon atom of the cytosine base, converting it into 5-methylcytosine (5mC).
- DNA methylation patterns are heritable, meaning they can be passed on from one cell generation to the next. During DNA replication, DNMT1 recognizes the hemimethylated DNA (one methylated and one unmethylated strand) and methylates the newly synthesized strand to maintain the methylation pattern.
- DNA methylation plays a crucial role in gene regulation. Generally, DNA methylation at gene promoters is associated with gene repression or silencing. The presence of methyl groups can hinder the binding of transcription factors and other regulatory proteins, leading to a less accessible chromatin structure and decreased gene expression.
- Besides gene promoters, DNA methylation can also occur in other genomic regions, such as gene bodies and intergenic regions. The functional consequences of DNA methylation in these regions are still being explored, but it is believed to influence alternative splicing, genomic stability, and long-range chromatin interactions.
- DNA methylation patterns are dynamically regulated throughout development, cellular differentiation, and in response to environmental cues. DNA demethylation can occur through active enzymatic processes or passively during DNA replication and repair.
In summary, DNA methylation is an epigenetic modification that involves the addition of a methyl group to cytosine residues in DNA. It regulates gene expression by influencing the accessibility of DNA to transcription factors and other regulatory proteins. The pattern of DNA methylation is heritable and dynamically regulated, playing a critical role in cellular processes and development.
Functions of DNA Methylation
DNA methylation is an epigenetic alteration that affects several biological processes. Here are some of the most important roles of DNA methylation:
- Regulation of gene expression: DNA methylation can regulate gene expression by reducing transcription factor binding to gene promoter regions. Methylation of promoter regions usually results in gene silencing, as it prevents the transcription machinery from accessing the DNA and hence reduces gene production.
- Developmental regulation: DNA methylation plays an important role in proper development and cellular differentiation. It aids in the establishment and maintenance of cell identity by designating certain genes for activation or suppression throughout development. During embryonic development and in response to environmental signals, methylation patterns can be dynamically changed.
- X-chromosome inactivation: DNA methylation is important in female X-chromosome inactivation. Each female cell has one of its two X chromosomes inactivated due to widespread DNA methylation. Because both males and females have one functioning X chromosome, this method guarantees dosage compensation.
- Genomic imprinting: DNA methylation is involved in genomic imprinting, an epigenetic mechanism that causes specific genes to be expressed based on their parental origin. During gametogenesis, methylation patterns are created and maintained in somatic cells, resulting in parent-of-origin-specific gene expression.
- Transposable element repression: Transposable elements (TEs) are DNA sequences that have the ability to move around the genome. DNA methylation suppresses TE activity, preventing transposition and consequent gene function disruption. Methylation serves as a defense mechanism to keep the DNA stable.
- DNA repair and stability: DNA methylation is involved in DNA repair mechanisms such as recognizing and repairing damaged DNA. It also helps to maintain genomic stability by limiting unauthorized recombination between repeated DNA segments.
- Association between sickness and cancer: Abnormal DNA methylation patterns have been related to a variety of disorders, including cancer. Changes in DNA methylation can result in gene expression dysregulation, which can contribute to cancer and disease development. In some malignancies, detecting DNA methylation patterns is also employed as a diagnostic and prognostic tool.
DNA Methylation Key Notes
- DNA methylation is an epigenetic mechanism in which a methyl (CH3) group is added to DNA.
- Methylation alters gene function and influences gene expression.
- The attachment of a methyl group to the 5-carbon of cytosine results in 5-methylcytosine (5-mC), which is the most common kind of DNA methylation.
- By projecting into the main groove of DNA, methyl groups in DNA hinder transcription.
- 5-methylcytosine is contained in roughly 1.5% of human genomic DNA.
- 5-mC is typically found in somatic cells in CpG sites, where a cytosine nucleotide is next to a guanine nucleotide.
- Normal somatic cell CpG islands are normally unmethylated, permitting gene expression.
- Methylation of a CpG island in the promoter region of a gene suppresses gene expression.
- DNA methyltransferases (DNMTs) are enzymes that add methyl groups to DNA.
- The creation and maintenance of DNA methylation patterns is mediated by three DNMTs (DNMT1, DNMT3a, and DNMT3b).
- DNMT1 keeps existing methylation patterns, whereas DNMT3a and DNMT3b create new methylation patterns.
- DNMT2 and DNMT3L may have specific DNA methylation roles.
- Diseased cells, such as cancer cells, may exhibit aberrant gene hypermethylation, which necessitates the collaboration of DNMTs 1 and 3b.
- The removal of a methyl group from DNA is known as DNA demethylation, and it is critical for gene reprogramming and disease processes.
- Demethylation can be passive, active, or a mix of the two.
- DNMT1 demethylates newly synthesised DNA strands during replication rounds, resulting in passive DNA demethylation.
- TET enzymes are involved in active DNA demethylation, which entails the elimination of 5-methylcytosine via cytosine base modification.
- TET1, TET2, and TET3 5-mC hydroxylases increase DNA demethylation by inhibiting DNA methyltransferase activity and converting 5-mC to 5-hmC, 5-fC, and 5-caC.
- TET proteins are involved in a variety of processes, including transcriptional activation and repression (TET1), tumour suppression (TET2), and DNA methylation reprogramming (TET3).
- 5-mC is a significant epigenetic alteration that influences phenotype and gene expression.
- DNA hypomethylation, defined as a decrease in global DNA methylation, can be used as a molecular marker in diseases such as cancer.
- The quantification of 5-mC concentration or global methylation can help in illness identification and analysis.
- The presence of the DNA demethylation intermediate 5-fC can be used to identify active DNA demethylation.
- Thymine DNA glycosylase (TDG) can remove 5-fC for subsequent base excision repair.
- Differentially methylated regions (DMRs) are DNA regions where the methylation state varies dramatically between samples.
- DMRs are identified via genome-wide methylation profiling, which reveals functional areas involved in gene transcriptional control.
- DMRs can be tissue, cell, or person specific.
- DMRs could be used as biomarkers or as targets for epigenetic treatment.
How does dna methylation affect gene expression?
DNA methylation is an epigenetic modification that plays a crucial role in regulating gene expression. Epigenetic modifications are changes to the DNA molecule that do not alter the underlying DNA sequence but can influence gene activity.
DNA methylation involves the addition of a methyl group (-CH3) to the cytosine base of a DNA molecule, typically occurring in a CpG dinucleotide context (where a cytosine is followed by a guanine). CpG sites are found throughout the genome, but they are often clustered in regions known as CpG islands, which are frequently located near the promoters of genes.
The presence of methyl groups on DNA can have two main effects on gene expression:
- Gene Silencing: Generally, DNA methylation at promoter regions of genes inhibits gene expression. Methylation interferes with the binding of transcription factors and other proteins required for the initiation of gene transcription. This prevents the recruitment of the transcriptional machinery necessary for the production of mRNA, leading to gene silencing or reduced gene expression. In other words, DNA methylation can act as a “switch” to turn genes off.
- Chromatin Structure Alteration: DNA methylation can also affect gene expression indirectly by modifying the structure of chromatin. Methylated DNA attracts proteins called methyl-binding domain (MBD) proteins, which recruit additional proteins, such as histone deacetylases, that modify the histones associated with DNA. These changes lead to a more condensed chromatin structure, known as heterochromatin. The condensed chromatin state makes the DNA less accessible to the transcriptional machinery, thus repressing gene expression.
It is important to note that DNA methylation patterns can be heritable, meaning they can be passed on from one generation to the next. This heritability plays a significant role in cellular differentiation and development, as well as in the maintenance of cell identity. However, DNA methylation patterns are also subject to dynamic changes throughout an individual’s life in response to various environmental factors and developmental cues, allowing for gene expression to be modulated in response to different conditions or stimuli.
Overall, DNA methylation provides a crucial mechanism for regulating gene expression and is involved in various biological processes, including development, cellular differentiation, and the response to environmental cues.
DNA methylation and histone acetylation
DNA methylation and histone acetylation are two major epigenetic changes that regulate gene expression.
The addition of a methyl group to the DNA molecule, specifically at cytosine residues in CpG dinucleotides, is known as DNA methylation. As previously stated, DNA methylation is commonly related with a decrease in gene expression. Methylated DNA can attract proteins known as methyl-binding domain proteins (MBDs), which can then recruit other proteins implicated in gene repression, resulting in chromatin compaction and gene transcription suppression. In general, higher levels of DNA methylation in a gene’s promoter region are linked to lower gene expression.
Histone acetylation, on the other hand, refers to the addition of an acetyl group to histone proteins, which are the proteins that wrap DNA to form a structure known as chromatin. Histone acetylation is commonly linked to enhanced gene expression. When histones are acetylated, the chromatin structure relaxes, enabling transcription factors and other proteins involved in gene transcription easier access to the DNA. Acetylated histones can also recruit transcriptional activation proteins.
DNA methylation and histone acetylation are frequently involved in the same epigenetic regulation process. High levels of DNA methylation in a gene’s promoter region are related with lower gene expression in many circumstances, whereas low levels of DNA methylation mixed with histone acetylation are associated with increased gene expression. The interaction of these epigenetic alterations, as well as other factors, influences the total amount of gene expression in a cell or organism.
It should be noted that the link between DNA methylation, histone acetylation, and gene expression can be complicated and context-dependent. Specific patterns and combinations of these alterations can differ between genes, cell types, and developmental stages, resulting in a wide range of gene expression consequences.
DNA methylation and cancer
DNA methylation is an epigenetic modification that plays a crucial role in normal cellular processes, including gene expression regulation and genome stability. Abnormal DNA methylation patterns have been observed in various diseases, including cancer. In cancer, alterations in DNA methylation can lead to the silencing of tumor suppressor genes or the activation of oncogenes, contributing to tumor development and progression.
There are two main types of DNA methylation changes observed in cancer:
- Hypermethylation: Hypermethylation refers to an increase in DNA methylation levels at specific regions, usually CpG islands, which are regions of DNA rich in cytosine and guanine nucleotides. CpG islands are often found in the promoter regions of genes and are associated with gene transcription regulation. Hypermethylation of CpG islands in gene promoters can lead to gene silencing, preventing the expression of tumor suppressor genes. Loss of tumor suppressor gene function can promote uncontrolled cell growth and division, contributing to cancer development.
- Hypomethylation: Hypomethylation refers to a decrease in DNA methylation levels, leading to genomic instability and aberrant gene expression. Global hypomethylation can occur in cancer and is associated with the activation of oncogenes, genomic instability, and chromosomal rearrangements. Hypomethylation may also lead to the reactivation of transposable elements (jumping genes), which can disrupt normal gene function and contribute to genomic instability.
The detection of DNA methylation changes in cancer has led to the development of promising biomarkers for cancer diagnosis, prognosis, and treatment response prediction. DNA methylation patterns can be analyzed through various techniques, such as bisulfite sequencing, methylation-specific PCR, and microarray-based methods. By assessing the DNA methylation status of specific genes or genomic regions, researchers and clinicians can gain insights into the molecular mechanisms underlying cancer and identify potential therapeutic targets.
Furthermore, targeting DNA methylation abnormalities in cancer has emerged as a therapeutic strategy. Drugs known as DNA methyltransferase inhibitors, such as 5-azacytidine and decitabine, can reverse DNA hypermethylation and reactivate silenced tumor suppressor genes. These drugs have been approved for the treatment of specific types of cancer, such as myelodysplastic syndrome, and are being investigated in clinical trials for other cancer types.
In summary, DNA methylation alterations play a significant role in cancer development by influencing gene expression and genomic stability. Understanding the specific changes in DNA methylation patterns in cancer can provide valuable insights into the underlying mechanisms and contribute to improved cancer diagnosis, prognosis, and the development of targeted therapies.
How is dna methylation used in dna repair?
DNA methylation plays a role in DNA repair through its involvement in the recognition and processing of DNA damage. Here are a few ways in which DNA methylation is implicated in DNA repair processes:
- Mismatch repair (MMR): MMR is a DNA repair pathway that corrects errors that occur during DNA replication, such as mismatched base pairs. DNA methylation can influence the recognition and repair of mismatches. Methylated DNA can recruit proteins that recognize and remove the mismatched bases, facilitating the repair process.
- Nucleotide excision repair (NER): NER is a DNA repair pathway that removes bulky DNA lesions, including UV-induced pyrimidine dimers and certain chemical adducts. DNA methylation patterns can affect the efficiency of NER by modulating the accessibility of repair proteins to the damaged sites. Methylated DNA can impede the binding of repair factors and hinder the repair process, while demethylation can promote repair efficiency.
- Base excision repair (BER): BER is a DNA repair pathway that corrects small, non-bulky lesions, such as damaged bases and single-strand breaks. DNA methylation can influence the recognition and processing of these lesions. For example, some DNA glycosylases, enzymes involved in BER, exhibit preference for specific DNA methylation states, enabling them to target and remove specific damaged bases.
- Double-strand break repair (DSBR): DNA double-strand breaks (DSBs) are severe DNA lesions that can lead to genomic instability and cell death if not properly repaired. DNA methylation can affect the choice of repair pathway for DSBs. Highly methylated regions of the genome tend to undergo repair through non-homologous end joining (NHEJ), while less methylated regions may preferentially utilize homologous recombination (HR) for repair.
These are just a few examples of how DNA methylation can influence DNA repair processes. The specific mechanisms and interplay between DNA methylation and DNA repair can vary depending on the context and the types of DNA lesions involved. Overall, DNA methylation can impact the efficiency and choice of DNA repair pathways, contributing to the maintenance of genome integrity.
Is dna methylation reversible?
Yes, DNA methylation is reversible. The addition and removal of methyl groups from DNA can occur through enzymatic processes, allowing for dynamic changes in DNA methylation patterns. This reversibility is important because it enables cells to regulate gene expression in response to environmental cues, developmental signals, and disease states.
The removal of DNA methylation marks, also known as DNA demethylation, can occur through two main mechanisms:
- Passive Demethylation: Passive demethylation occurs through DNA replication. When a methylated DNA strand is replicated, the newly synthesized strand is initially unmethylated. Over subsequent rounds of replication, if DNA methyltransferase enzymes are not present or active, the methylation marks may not be restored, leading to a gradual loss of DNA methylation over multiple cell divisions.
- Active Demethylation: Active demethylation involves enzymatic processes that actively remove the methyl groups from DNA. One well-known pathway of active demethylation involves ten-eleven translocation (TET) enzymes. TET enzymes can oxidize methylated cytosine residues to generate intermediates that are recognized and repaired by the cell’s DNA repair machinery, ultimately leading to the removal of the methyl group.
Additionally, DNA methyltransferase inhibitors, such as 5-azacytidine and decitabine, can be used to induce DNA demethylation by preventing the addition of methyl groups during DNA replication. These inhibitors are used in clinical settings for certain diseases, including myelodysplastic syndrome and acute myeloid leukemia.
It is important to note that while DNA methylation is reversible, the restoration of a specific methylation pattern can vary depending on the context and the dynamics of the underlying regulatory mechanisms.
How to measure dna methylation?
DNA methylation can be measured using various techniques, including:
- Bisulfite Sequencing: This is the gold standard method for DNA methylation analysis. Bisulfite treatment converts unmethylated cytosines to uracils while leaving methylated cytosines unchanged. After bisulfite treatment, DNA sequencing can be performed to determine the methylation status of specific regions of the genome.
- Methylation-Specific PCR (MSP): This method amplifies DNA regions of interest after bisulfite treatment using primers specific to either methylated or unmethylated DNA sequences. MSP can provide qualitative information about the presence or absence of methylation at specific sites.
- Combined Bisulfite Restriction Analysis (COBRA): COBRA combines bisulfite treatment with enzymatic digestion using methylation-sensitive restriction enzymes. The digested DNA fragments are then analyzed by PCR or sequencing. COBRA allows for semi-quantitative assessment of DNA methylation.
- Methylation-Sensitive High-Resolution Melting (MS-HRM): This technique utilizes high-resolution melting analysis after bisulfite treatment to differentiate methylated and unmethylated DNA based on their melting profiles. MS-HRM is a sensitive and cost-effective method for screening DNA methylation changes.
- Pyrosequencing: Pyrosequencing is a quantitative DNA sequencing-based method that can measure DNA methylation at multiple CpG sites. It provides detailed information about the degree of methylation at individual CpG dinucleotides.
- DNA Methylation Microarrays: Microarray platforms, such as Illumina Infinium Methylation BeadChip, can analyze DNA methylation patterns across the genome. These arrays contain probes specific to methylated and unmethylated DNA regions, and the intensity of hybridization can be used to determine the methylation status.
- Next-Generation Sequencing (NGS): NGS technologies, like whole-genome bisulfite sequencing (WGBS) or reduced representation bisulfite sequencing (RRBS), provide comprehensive and high-resolution DNA methylation maps. These methods involve bisulfite treatment, followed by high-throughput sequencing of the converted DNA.
Each technique has its advantages and limitations, and the choice of method depends on the research goals, sample size, and available resources.
DNA methylation vs acetylation/DNA acetylation vs methylation
DNA methylation and DNA acetylation are two distinct epigenetic modifications that play important roles in gene regulation and chromatin structure. While they both involve chemical modifications of DNA, they have different effects on gene expression.
DNA Methylation: DNA methylation refers to the addition of a methyl group (-CH3) to the cytosine base of DNA, typically occurring at cytosine-guanine dinucleotides (CpG sites). DNA methylation is often associated with gene silencing and transcriptional repression. When CpG sites in the promoter region of a gene are methylated, it can inhibit the binding of transcription factors, leading to reduced gene expression. DNA methylation is involved in various biological processes, including embryonic development, X-chromosome inactivation, and the maintenance of genomic stability.
DNA Acetylation: DNA acetylation involves the addition of an acetyl group (-COCH3) to the DNA molecule. Acetylation occurs on the histone proteins around which DNA is wrapped, rather than directly on the DNA itself. Histones are proteins that help package DNA into a condensed structure called chromatin. Acetylation of histones is associated with gene activation and transcriptional activity. Acetyl groups neutralize the positive charge of histones, reducing the interaction between histones and DNA. This relaxed chromatin structure allows for easier access of transcription factors and other regulatory proteins to the DNA, promoting gene expression.
Differences:
- Location: DNA methylation occurs directly on the DNA molecule at CpG sites, while DNA acetylation occurs on the histone proteins around which DNA is wrapped.
- Effect on Gene Expression: DNA methylation is generally associated with gene repression and silencing, whereas DNA acetylation is associated with gene activation and increased expression.
- Mechanism: DNA methylation primarily affects gene expression by inhibiting the binding of transcription factors to DNA, while DNA acetylation affects gene expression by modifying the chromatin structure and making the DNA more accessible to transcription factors.
- Stability: DNA methylation is relatively stable and can be maintained through cell divisions, providing a long-term epigenetic mark. In contrast, DNA acetylation is more dynamic and reversible, allowing for more rapid changes in gene expression.
Both DNA methylation and DNA acetylation are essential for normal cellular function and development, and disruptions in these epigenetic modifications can contribute to various diseases, including cancer and neurological disorders.
DNA Methylation and Disease
DNA methylation is an epigenetic modification that plays a crucial role in the regulation of gene expression. It involves the addition of a methyl group to the DNA molecule, specifically at cytosine residues in CpG dinucleotides. DNA methylation patterns are essential for normal development and cellular differentiation, and alterations in these patterns have been implicated in various diseases.
Here are some ways in which DNA methylation is associated with disease:
- Cancer: Abnormal DNA methylation patterns are commonly observed in cancer cells. Two major types of alterations are seen: global hypomethylation and localized hypermethylation. Global hypomethylation refers to the overall decrease in DNA methylation levels, while localized hypermethylation involves the aberrant methylation of specific CpG islands within the promoter regions of tumor suppressor genes. These changes can lead to the silencing of tumor suppressor genes or the activation of oncogenes, contributing to tumor development and progression.
- Neurological disorders: DNA methylation has been implicated in various neurological disorders, including neurodevelopmental disorders like autism spectrum disorders (ASD) and Rett syndrome. Studies have shown altered DNA methylation patterns in the brains of individuals with these conditions. DNA methylation changes in specific genes can affect neuronal development, synaptic function, and neurotransmitter signaling, contributing to the pathogenesis of these disorders.
- Cardiovascular disease: DNA methylation alterations have been observed in cardiovascular diseases such as atherosclerosis, hypertension, and heart failure. Methylation changes in genes related to inflammation, lipid metabolism, and endothelial function can impact the development and progression of these conditions. DNA methylation patterns can also serve as biomarkers for predicting cardiovascular disease risk.
- Autoimmune disorders: Aberrant DNA methylation has been implicated in autoimmune diseases like rheumatoid arthritis, systemic lupus erythematosus, and multiple sclerosis. Altered DNA methylation patterns can affect immune cell function, leading to dysregulated immune responses and self-reactivity. Methylation changes in genes involved in immune regulation and inflammation can contribute to the development and progression of autoimmune disorders.
- Aging: DNA methylation patterns undergo changes with age, commonly referred to as epigenetic aging. These age-associated methylation changes have been linked to age-related diseases such as cardiovascular disease, neurodegenerative disorders, and cancer. Epigenetic clocks, which estimate biological age based on DNA methylation patterns, have been developed and show promise as predictors of age-related disease risk and mortality.
FAQ
What is DNA methylation?
DNA methylation is a chemical modification process where a methyl group is added to a cytosine residue in DNA, often occurring at specific sites called CpG dinucleotides.
What is the role of DNA methylation?
DNA methylation plays a crucial role in gene regulation by affecting the binding of transcription factors to DNA, thereby influencing gene expression. It is involved in various biological processes, including development, genomic imprinting, and X-chromosome inactivation.
How is DNA methylation carried out?
DNA methylation is carried out by enzymes called DNA methyltransferases (DNMTs). DNMT3A and DNMT3B are responsible for de novo methylation, while DNMT1 maintains existing methylation patterns during DNA replication.
Can DNA methylation patterns change?
Yes, DNA methylation patterns can change in response to various factors, such as environmental cues, aging, and disease. This flexibility allows for the regulation of gene expression and adaptation to different cellular contexts.
Is DNA methylation heritable?
DNA methylation patterns can be heritable, meaning they can be passed on from one generation to the next. This inheritance pattern is important for maintaining cellular identity and regulating gene expression during development.
What is the difference between de novo methylation and maintenance methylation?
De novo methylation refers to the addition of methyl groups to previously unmethylated DNA, establishing new methylation patterns. Maintenance methylation, on the other hand, preserves existing methylation patterns during DNA replication to ensure their faithful transmission to daughter cells.
Can DNA methylation be reversed?
Yes, DNA methylation can be reversed through a process called DNA demethylation. Demethylation can occur through passive mechanisms during cell division or active mechanisms involving enzymatic reactions that remove the methyl groups.
What are the consequences of abnormal DNA methylation?
Abnormal DNA methylation patterns are associated with various diseases, including cancer, neurological disorders, and developmental abnormalities. Dysregulation of DNA methylation can disrupt normal gene expression patterns and cellular processes.
How is DNA methylation studied?
Scientists study DNA methylation using various techniques, including bisulfite sequencing, methylation-specific PCR (MSP), and methylation microarrays. These methods allow researchers to analyze DNA methylation patterns at specific genomic regions or on a genome-wide scale.
Can DNA methylation be targeted for therapeutic purposes?
The ability to modify DNA methylation patterns holds potential for therapeutic interventions. Epigenetic drugs, such as DNA methyltransferase inhibitors, are being explored as potential treatments for certain diseases, particularly cancer, by reprogramming aberrant DNA methylation patterns.
dna methylation and histone acetylation are examples of _
DNA methylation and histone acetylation are examples of epigenetic modifications. Epigenetic modifications are heritable changes in gene expression that do not involve alterations to the DNA sequence itself. Instead, they involve modifications to DNA or histone proteins that can influence gene activity.
DNA methylation refers to the addition of a methyl group to the DNA molecule, usually at cytosine residues in a CpG dinucleotide context. This modification can result in gene silencing by inhibiting the binding of transcription factors and other regulatory proteins to the DNA, thus reducing gene expression.
Histone acetylation, on the other hand, involves the addition of an acetyl group to specific lysine residues on histone proteins. This modification generally leads to relaxed chromatin structure and increased accessibility of the DNA to transcription factors and other regulatory proteins. As a result, gene expression is often enhanced.
Both DNA methylation and histone acetylation play important roles in gene regulation, development, and various cellular processes. They are key mechanisms by which cells can modify gene expression patterns in response to environmental cues and developmental signals.
dna methylation leads to methyl groups being added to cpg, which in turn:
DNA methylation leads to methyl groups being added to CpG dinucleotides, which can have several effects on gene expression and cellular function:
Gene Silencing: DNA methylation at CpG islands located near gene promoters is often associated with gene silencing or reduced gene expression. The presence of methyl groups can interfere with the binding of transcription factors and other regulatory proteins to the DNA, inhibiting gene transcription.
Stable Gene Repression: DNA methylation can provide long-term stability to gene repression. Once methylated, CpG sites tend to be maintained across cell divisions and can transmit the silenced state to daughter cells.
X-Chromosome Inactivation: DNA methylation plays a critical role in X-chromosome inactivation, a process that equalizes gene expression between males and females during early embryonic development. One of the X chromosomes in females becomes extensively methylated, leading to its inactivation.
Genomic Imprinting: DNA methylation also contributes to genomic imprinting, a phenomenon where specific genes are differentially expressed depending on their parental origin. Imprinted genes have methyl marks that are inherited from one parent and influence their expression pattern.
It’s important to note that DNA methylation can be dynamic and reversible, meaning that changes in methylation patterns can occur during development, in response to environmental factors, or in certain diseases.
Does dna methylation increase gene expression?
No, DNA methylation typically leads to a decrease in gene expression, although the relationship between DNA methylation and gene expression is complex and context-dependent. DNA methylation is an epigenetic modification that involves the addition of a methyl group to the DNA molecule, specifically to cytosine residues in CpG dinucleotides. CpG dinucleotides are regions where a cytosine nucleotide is followed by a guanine nucleotide in the DNA sequence.
High levels of DNA methylation in the promoter region of a gene, which is the region responsible for regulating gene expression, are often associated with reduced gene expression. DNA methylation can inhibit gene expression by preventing the binding of transcription factors, which are proteins that regulate the transcription of genes, to the promoter region. Additionally, methylated DNA can attract proteins that further repress gene expression.
However, it’s important to note that the relationship between DNA methylation and gene expression is not always straightforward. In some cases, DNA methylation may be associated with increased gene expression. For example, DNA methylation patterns can vary across different tissues and developmental stages, and the relationship between DNA methylation and gene expression can be gene-specific.
Overall, while DNA methylation is generally associated with decreased gene expression, the specific effects can vary depending on the gene and the context in which it occurs.
is DNA methylation good or bad?
DNA methylation itself is neither inherently good nor bad. It is a natural and essential process that regulates gene expression and contributes to normal cellular function. DNA methylation is crucial for normal development, cellular differentiation, and the maintenance of genome stability.
However, abnormalities in DNA methylation patterns can have negative consequences and are associated with various diseases, including cancer. In cancer, DNA methylation changes can lead to the silencing of tumor suppressor genes, which are important for controlling cell growth and preventing tumor formation. Additionally, DNA methylation abnormalities can activate oncogenes, which promote uncontrolled cell growth and division.
So, while DNA methylation is a normal and necessary process, its dysregulation in the context of diseases such as cancer can have detrimental effects. Understanding and targeting these aberrant DNA methylation patterns can provide opportunities for diagnosis, prognosis, and the development of therapeutic interventions.
References
- Moore, L., Le, T. & Fan, G. DNA Methylation and Its Basic Function. Neuropsychopharmacol 38, 23–38 (2013). https://doi.org/10.1038/npp.2012.112
- Jin B, Li Y, Robertson KD. DNA methylation: superior or subordinate in the epigenetic hierarchy? Genes Cancer. 2011 Jun;2(6):607-17. doi: 10.1177/1947601910393957. PMID: 21941617; PMCID: PMC3174260.
- https://www.news-medical.net/life-sciences/What-is-DNA-Methylation.aspx
- https://www.whatisepigenetics.com/dna-methylation/
- https://www.sciencedirect.com/topics/neuroscience/dna-methylation
