Table of Contents
The DNA microarray technology is among the most effective technology that can offer an extremely high-throughput and precise overview of the whole transcriptome and genome, which lets scientists understand the molecular processes that underlie natural and malfunctioning biological processes. Microarray technology may accelerate the process of screening thousands of protein and DNA samples at once.
What is dna microarray?
- DNA Microarray was first introduced as a technique in E.M. Southern’s publication of 1970. Today, it has become an efficient technology platform to finding biomarkers that are valid by combining transcriptomic samples against nearly genome-wide gene sets simultaneously.
- Spotted DNA microarrays have evolved into an extremely valuable technology that was developed in the middle of the past decade, when an initial DNA microarray created in the lab of Schena et al.
- A typical microarray study involves the mixing of an mRNA-containing molecular in the template DNA the source of its origin.
- A variety of DNA samples are used to build an array. It is the amount of mRNA attached to each spot in the array is a sign of the level of expression of diverse genes. The number could be in the thousands.
- All data is collected and a profile created to measure the expression of the gene within the cell.
- DNA microarray (also popularly referred to as DNA chip, gene chip as well as biochip) is an array of tiny DNA spots that are connected to a solid surface.
- In the DNA chip technology, DNA molecules that are single-stranded are stuck to a surface using biochemical analysis.
- The DNA molecules come from cDNA sequences found in an organism, referred to as cDNA microarray , or photolithography to synthesize short sequences of oligonucleotides.
- Researchers use DNA microarrays to monitor the levels of expression of a number of genes in a single test or to genotype several regions of the genome.
- Every DNA spot has picomoles (10″12 moles) of a particular DNA sequence, referred to as probes (or reporters). They can be short sections of a gene or any other DNA-based element used to create hybrids of the cDNA or cRNA samples (called target) under conditions of high-stringency.
- Probe-target hybridization is usually detected and quantified by detection of flurophore- , silver-, or chemiluminescence-labeled targets to determine relative abundance of nucleic acid sequences in the target.
- The most recent generation of biosensors is biochips, which are created by using DNA probes.
- DNA microarrays are typically made from glass or silicon which is where the DNA is fixed.
- The expression of tens and thousands genes can be analyzed using the DNA microarray.
- cDNA-based microarray and oligonucleotide-based microarray are the two different types of microarray.
- To perform high-throughput and large-scale genome analyses, the most effective and well-established method is utilized, which is known as DNA microarray.
- The technology was initially designed to determine the level of transcription in the RNA transcripts extracted from a variety of genes.
- The research on DNA microarrays isn’t only limited to gene expression. It can be utilized to identify single nucleotide polymorphisms (SNPs) or patterns that have been methylated and alternative RNA splicing modification in the copy number of a gene and pathogen detection.
Definition of DNA microarray
The DNA microarray is a type of tool that can be used to find out if the DNA of an person has a mutation in genes such as BRCA1 or BRCA2. The chip is comprised of a tiny glass plate enclosed in plastic. Certain companies make microarrays by using techniques similar to those that are used to manufacture microchips for computers. On the outside every chip is made up of thousands of single-stranded DNA sequences that are synthetic all of which make up the gene that is normal as well as to variations (mutations) of the gene discovered within the human population.
Principle of DNA microarray
- The principle of DNA microarrays is based on the hybridization between nucleic acid strands. Complementary nucleic acid sequences can specifically pair with each other by forming hydrogen bonds between complementary nucleotide base pairs. In DNA microarray technology, samples are labeled with fluorescent dyes, and at least two samples are hybridized to a chip.
- During hybridization, the complementary nucleic acid sequences between the sample and the probe attached to the chip form hydrogen bonds. Non-specific bonding sequences remain unattached and are washed out during the washing step of the process. The fluorescently labeled target sequences that bind to the probe generate a signal.
- The strength of the signal depends on various factors such as the hybridization conditions (e.g., temperature) and washing after hybridization. The total strength of the signal depends on the amount of target sample present. By utilizing this technology, it is possible to screen the presence of one genomic or cDNA sequence among 100,000 or more sequences in a single hybridization.
- DNA microarray technology was derived from Southern blotting, which involves attaching fragmented DNA to a substrate and probing it with a known DNA sequence. In DNA microarrays, the unknown DNA sample is referred to as the target, while the known DNA sequence is called the probe. The samples are labeled with fluorescent dyes, and at least two samples are hybridized to the chip.
- The strength of the hybridization between the target and probe is determined by the number of complementary base pairs in the nucleotide sequence. After washing off non-specific bonding sequences, only strongly paired strands will remain hybridized. The fluorescently labeled target sequences that pair with the probe release a signal, which depends on the strength of the hybridization detected by the number of paired bases, hybridization conditions, and washing after hybridization.
- DNA microarrays employ relative quantization, where the comparison of the same character is done under two different conditions, and the identification of that character is known by its position on the microarray. After hybridization is complete, the surface of the chip can be examined qualitatively and quantitatively using techniques such as autoradiography, laser scanning, fluorescence detection devices, or enzyme detection systems.
- Overall, DNA microarrays enable the screening of the presence of specific genomic or cDNA sequences among a large number of sequences in a single hybridization, providing valuable insights into gene expression, genotyping, and other genetic analyses.
Types of DNA microarray
There are four kinds of DNA microarrays:
- Oligo DNA microarray: It is made up of the oligonucleotides with a length of 20-50 nucleotides. Oligonucleotides are made directly onto the slides. A single color hybridization is used to make each probe. It is very specific but low sensitivity.
- cDNA microarray: It is often referred to as a microarray that is spotted. It comprises DNA fragments with any size (500bp-1kb) or 20-100 nt oligos are glued to glassslides. It makes use of two colors hybridization for each probe.
- BAC Microarray: It is made using the template, which is amplified through polymerase chain reaction to form the probe.
- SNA Microarray: It’s utilized to determine polymorphisms within the population.
1. cDNA based microarrays or Glass cDNA microarrays
This kind of array contains cDNA fragments that range from 600 to 2400 nucleotides long. In order to make the cDNA microarray every probes have to be picked independently and produced using the process of PCR or cloning. Then , all DNA probes are put on the slide.
- cDNA microarray can also be referred to as an spotted microarray. This is among the first widely accessible array platforms.
- Glass DNA microarrays were the first form of DNA microarrays that were developed. It was developed by Patrick Brown and his colleagues at Stanford University and is produced with a robot that deposits (spots) one nanoliter of DNA (50-150 um in diameter) onto a microscope coated glass slide in serial order , with an approximate distance of 200-250 millimeters from one another one gene.
- The moderately sized glass microarrays for cDNA contain approximately 10.000 spots, or even more an expanse of 3.6 cm2.
- Like the name implies, glass cDNA microarrays are made from specially designed glass slides that have desirable physicochemical properties e.g. outstanding chemical resistance to solvents, excellent mechanical stability (increased temperature strain points) and low intrinsic fluorescence characteristics.
- To create an entire genome ceramic DNA microarray it is necessary to follow a series of steps that are repeated with each step needing an appropriate and well-planned approach.
Characteristics of cDNA based microarrays
- cDNA microarray is also known as an spotted microarray. This is among the first broadly and widely accessible array platforms.
- To make chips cDNA can be used.
- It is employed for amplifying of cDNA.
- CDNA-based microarrays are a very high-throughput method.
- The microarray technique permits the qualitative analysis of transcripted RNAs of known and unknown genes . It can also be described as a multi-dimensional RNA expression assay.
- Glass cDNA microarrays are inexpensive and does not need special equipment to hybridize, the sensitivity to detect is increased because of the longer targeted sequences. Also, to create a DNA sequence primary sequence information isn’t required.
Steps of Glass cDNA microarrays
- The first step to make a cDNA microarray made of glass is selecting the appropriate material to be spotted on the glass microscope surface e.g. the genes from public databases/repositories or institutional sources.
- The next step is the extraction and the purification of DNA sequences that represent the gene of interest.
- In the process of preparation, PCR is used to amplify the DNA of the libraries of interest by using universal primer or gene-specific primers. The quality of the DNA fragments that represent genes of interest is usually verified by sequencing or on agarose gels to simultaneously determine the concentration of DNA. This is an important step because all the DNA fragments should be of similar concentration/molarity and size, to achieve similar reaction kinetics for all hybridisations.
- The third step is spotting DNA solution onto chemically modified glass slides usually with poly(L-lysine) or other cross-linking chemical coating materials such as polyethyleneimine polymer p-aminophenyl trimethoxysilane/diazotization chemistry and dendrimeric structure.
- It is the substrates that are coated on the outside of the slide which determines how DNA will be held on the slide’s surface e.g. covalent or not.
- In the course of poly(L-lysine) the positively charged phosphate groups within the DNA molecule, create an ionic connection with the positively charged aminederivatised surface.
- This spotting procedure is carried out by a contact printing process using precise robotic pins, or any other similar technologies for delivering information, such as inkjet printing.
- The final stage of producing glass DNA microarrays involves the post-print processing process, which involves drying process of the DNA slide for a night at room temperature and use of UV cross-linking in order to hinder the further DNA binding and also to reduce the background signal after the hybridisation of a target labelled.
Advantages of cDNA microarrays
- The advantages that come with Glass cDNA microarrays are their cost-effectiveness and lower price.
- Its accessibility , which does not require any special equipment, which means that hybridisation doesn’t require special equipment. Data capture can be done with equipment that is frequently already in the lab and a variety of it is possible to design the experiment in a way that is influenced by the goals of science of the research.
- Additionally, Glass cDNA microarrays also offer increased sensitivity to detection due to the longer targeted sequences ( 2.25 kbp).
Disadvantages of cDNA microarrays
- Despite their popularity of glass microarrays, they come with a few drawbacks, like the need for labor intensive for the synthesis, purification and storing DNA solutions before the microarray’s fabrication.
- Additionally, there are more printing equipment is needed, which makes microarrays more costly.
- Additionally, during microarray tests in the lab the homologies in sequences between clones that represent different close relatives of the same family can result in the failure of specifically identify individual genes and may instead hybridize to a spot(s) that are designed to identify the transcript of an entirely different gene. This is known as cross-hybridisation.
2. Oligonucleotide based microarray or in situIn situ oligonucleotide array
- The In-Situ (on chip) Oligonucleotide array format is an advanced microarray platform technology, which is manufactured by using the technique of chemical synthesis in situ that was invented by Stephen Fodor et al. (1991).
- However, the market leading company in in-situ microarrays of oligonucleotides (Affymetrix) has also developed this kind of technology for the production of the so-called
- GeneChips is a reference to its high-density DNA arrays made of oligonucleotides.
- The Commercial versions of Affymetrix GeneChips can accommodate up to 500,000 probes/sites within the 1.28cm2 area. Due to the extremely high amount of information (genes) they are being used extensively in the detection of hybridisations and analysis of polymorphisms and mutations like single nucleotide polymorphisms and disease-related genetic mutation study (“genotyping”) and a broad array of other applications, such as study of gene expression, to just some.
- The fundamental tenets of making Affymetrix’s GeneChips is using photolithography and combinatorial chemical to create small single DNA strands on 5 inch round quartz chips.
- Contrary to glass cDNA and cDNA on glass, the chips’ genes are designed on the basis of sequence information by itself followed by an industrial chip synthesiser, sequences are directly synthesized onto an area of 5 inch square quartz wafer in pre-selected location.
Characteristics of Oligonucleotide based microarray
- The microarray of Oligonucleotide hybridizes using only one sample, and provides the exact levels of expression of the samples since it is extremely specific.
- Through this technique the photolithographic technique is us, the technique is employed to create high-density microarray chips . The array is created by synthesizing single-stranded oligonucleotides the field.
- The storage and collection of the cloned DNA as well as products of PCR are not required for this process.
- Expression and analysis of genes are only limited by this method since it requires a vast quantity of biological materials.
- It is extremely precise rapid, reliable, and fast.
- There are a variety of dsDNA probes are simpler to create, but despite this they must be designed with care so that all probes have similar melting temperatures , and also eliminate palindromic sequences.
- Because of its smaller size of the probe cross-link could result in a substantial loss due to washing.
- The modified 5′-3 ends of coated slides assist in coupling probes to the microarray’s surface, which provides functional groups, such as aldehyde or epoxy.
Steps of in situIn situ oligonucleotide array format
- The manufacturing process for Affymetrix’s GeneChips with DNA photolithography starts with the derivatization process of the solid support. Typically, it is quartz, with the covalent linker molecule, which is then that is then protected by a photolabile group.
- This is accomplished by cleaning the quartz to ensure uniform hydroxylation over the surface before setting it into a silane bath that reacts with hydroxyl groups in the quartz to form an interconnected matrix of molecules.
- The in-situ synthesis of Oligonucleotides is performed simultaneously, leading to the successive additions of A, C, G and T nucleotides onto the appropriate sequences of genes within the array. In each stage of the synthesizing process, the chains of oligonucleotides that, for instance, require adenine at the following position are uncovered by light at the proper places by masks.
- Its quartz (chip) can then filled with a solution of activated adenine nucleotides, with an ejectable protection group that are then connected to the deprotected positions.
- Adenine residues that are uncoupled are cleaned off and a new mask is put on to perform the deprotection process that follows the nucleotide.
- In the end, repeating the process 70 times, using 70 different masks, permits the synthesis of all of tens of thousands of 25-mer oligonucleotides that can be synthesised in.
Advantages of Oligonucleotide based microarray or in situ oligonucleotide array
- Benefits of using the in-situ oligonucleotide array include accuracy, speed and precision.
- The speed of making the array is the main benefit because, placing the DNA on the chip is as simple as requiring the DNA sequence in question be known. Therefore, there is no need to spend time handling CDNA resources like the preparation and precise identification of the bacterial clones the PCR products or cDNAs which reduces the risk of contamination and mixing up.
- However, before making the array, information about the DNA sequence of the organism is necessary in order to create the oligonucleotide sets and, if this isn’t available, alternative ways of printing genetic material isolated could be considered.
- Other benefits of the in-situ format for oligonucleotide arrays are its high specificity and repeatability. Both of these advantages are due to the manner in which the sequences of oligonucleotides to be put on the chips are created and the inclusion of multiple short sequence(s) which represent the distinctive genetic sequence.
- In the case of designing an oligonucleotide-based sequence for a particular gene the sequences are designed to be totally compatible with the target gene sequence in addition, an additional sequence partner is created that is the same except for one base mismatch at the middle. This strategy of mismatching sequences, in conjunction with the usage of more than one sequence(s) to each gene, improves the accuracy and allows for the identification and minimize the impact of non-specific hybridisation as well as background signals. This method also permits the removal of cross-hybridisation signal and the differentiation between signals that are real and non-specific.
Disadvantages of Oligonucleotide based microarray or in situ oligonucleotide array format
- There are a number of disadvantages to the in-situ oligonucleotide array design, including limitations in terms of cost and the flexibility.
- First of all, in-situ designs for oligonucleotides typically require expensive equipment e.g. to perform an hybridization, staining labels, washing, and quantitation.
- Additionally, pre-fabricated in-situ oligonucleotide arrays (GeneChips) are not cheap; however, there has been a reduction in costs as the market for microarrays has grown.
- Thirdly, even though short-sequences that are used on the array offer the highest specificity, they might have lower sensitivity or binding compared to glass microarrays containing cDNA. This low sensitivity is compensated by the use of multiple probes.
- The In-Situ oligonucleotide format is also less flexible, even though it isn’t the case for the design of the array. There are instances where the manufacturing of arrays, hybridisation , and detection equipment is restricted to centralized manufacturing facilities which limits the flexibility of researchers.
- The cost and time required to make the in-situ oligonucleotide array makes it prohibitive for the average lab to synthesize the chips itself.
Classification of microarray based on the types of probes used
Based on the kinds of probes that are used microarrays can be classified into twelve distinct kinds:
1. DNA microarrays
- DNA microarray is also referred to as DNA chip, gene chip chip, biochip.
- It can either measure DNA or makes use of DNA as part the detection process.
- There are four kinds of DNA microarrays: cDNA microarrays, oligo DNA microarrays BAC microarrays, and SNP microarrays.
2. MMChips
- MMchip permits the integration of analysis of cross-platform data and between-laboratory data.
- It studies the interactions between protein and DNA.
- ChIP chip (Chromatin immunoprecipitation (ChIP) followed by array hybridization) and
- ChIP-seq (ChIP followed by massively parallel sequencing) are two methods employed.
3. Protein microarrays
- It serves as a platform for the analysis from hundreds of millions of proteins, in a parallel fashion.
- Protein microarrays are of three kinds, and they are analytical microarrays of proteins, functional protein microarrays, and reverse-phase microarrays of proteins.
4. Peptide microarrays
- These kinds of arrays are utilized for detailed analysis or for optimizing protein-protein interactions.
- It aids in the recognition of antibodies in the process of screening proteomes.
5. Tissue microarrays
- Paraffin blocks of tissue that are created by segregating cylindrical tissue cores of various donors before encapsulating it into one microarray.
- It is most commonly used in pathology.
6. Cellular microarrays
- They are also called transfection microarrays or living-cell-microarrays, and are used for screening large-scale chemical and genomic libraries and systematically investigating the local cellular microenvironment.
7. Chemical compound microarrays
- This is used to screen drugs and for drug discovery.
- This microarray can be used to determine and evaluate small molecules, which is why it’s more efficient than the other methods utilized in the pharmaceutical industry.
8. Antibody microarrays
- They can also be called antibodies arrays or antibody chips.
- They are specifically designed microarrays for proteins. They include a variety of antibodies that are captured and placed on the microscope slide.
- They are used to detect antigens.
9. Carbohydrate arrays
- They can also be referred to as glycoarrays.
- Carbohydrate arrays can be used for screening proteomes with carbohydrate bound.
- They are also used to calculate affinities for protein binding and in the automization of solid-support synthesis for glycocans.
10. Phenotype microarrays
- PMs or Phenotype microarrays are used for the development of drugs.
- They can quantitatively assess thousands of cellular characteristics all simultaneously.
- It is also utilized in functional genomics as well as toxicological tests.
11. Reverse phase protein microarrays
- These are microarrays of serum or lysates.
- They are mostly utilized in clinical trials particularly within the cancer field research, they can also be used for pharmaceutical purposes.
- In certain instances they could also be utilized to study biomarkers.
12. Interferometric reflectance imaging sensor or IRIS
- IRIS is a biosensor utilized to examine protein-protein DNA, protein-DNA, as well as DNA-DNA interactions.
- It doesn’t make use of fluorescent labels.
- It is constructed of Si/SiO2 films that are prepared through robotic spot-spotting.
Requirements of DNA microarray
To create a DNA microarray, several requirements must be met, including:
- DNA Chip: The DNA chip, also known as the microarray, is a solid surface (usually a glass slide or silicon wafer) that contains an array of DNA probes immobilized at specific locations. The chip should be designed to accommodate a sufficient number of probes to capture the desired genomic or cDNA sequences.
- Target Sample: The target sample refers to the DNA or RNA that is being analyzed. It should be fluorescently labeled using fluorescent dyes or other labeling methods. The labeling enables the detection and visualization of the hybridization between the target sample and the DNA probes on the chip.
- Fluorescent Dyes: Fluorescent dyes are used to label the target samples. These dyes emit fluorescence when excited by specific wavelengths of light. Different dyes can be used to label different samples, allowing for multiplexing and simultaneous detection of multiple targets on the microarray.
- Probes: Probes are short DNA or RNA sequences that are complementary to the target sequences of interest. They are attached to the surface of the DNA chip at specific locations. The probes should be carefully designed to ensure specificity and sensitivity in hybridizing with the target samples.
- Scanner: A scanner is required to read and capture the fluorescent signals emitted by the labeled target samples bound to the DNA probes on the chip. The scanner detects the intensity of fluorescence at each probe location and converts it into digital data for subsequent analysis.
DNA microarray steps
The process of reaction for DNA microarray is carried out in a series of phases:
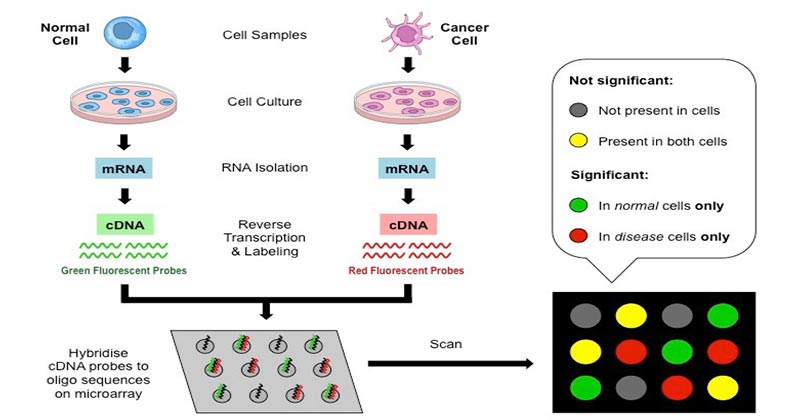
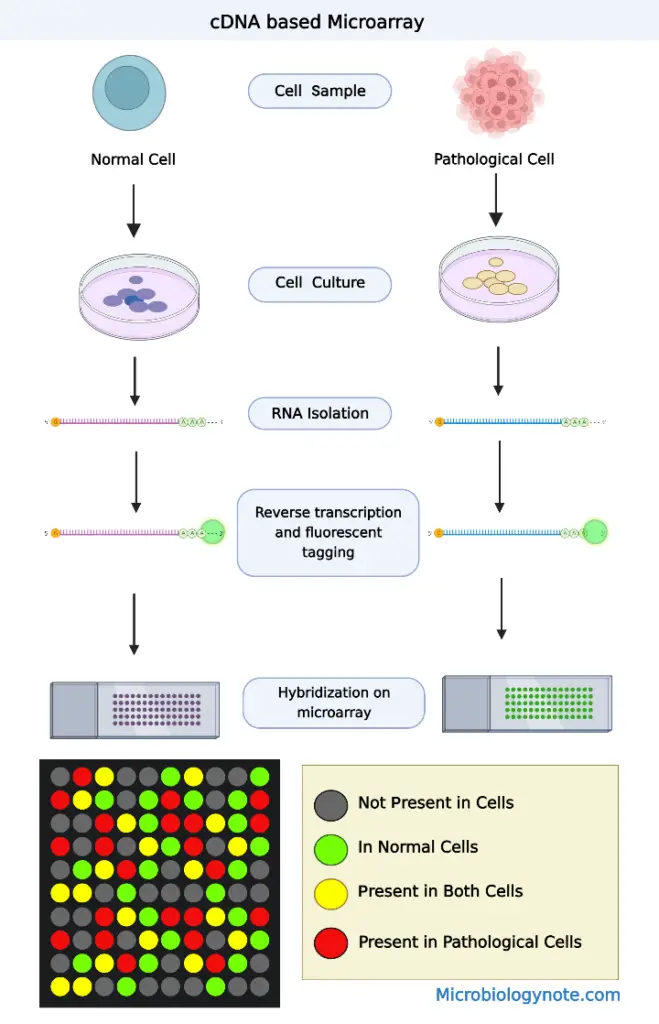
1. Collection of samples
- The sample could be a tissue or cell of the animal we would like to conduct our study on.
- Two kinds of samples are taken from healthy cells and infected ones, to compare them and get the outcomes.
2. Isolation of mRNA
- The RNA is extracted from the sample by using an instrument or solvent such as phenol-chloroform.
- The RNA extracted from the extract MRNA is split, leaving behind rRNA and TRNA.
- Since mRNA is an A-polymer tail the poly-T-tails of column beads can be used to bind mRNA.
- After extraction the column is washed with a buffer to remove mRNA from beads.
3. Creation of labeled cDNA
- In order to create cDNA (complementary DNA Strand) reverse transcription of the mRNA performed.
- The two samples are combined with various fluorescent dyes to create fluorescent CDNA Strands. This allows for the identification of the categories of samples of cDNAs.
4. Hybridization
- The cDNAs with labels from both samples are placed into the DNA microarray to ensure that each cDNA will be connected to the complementary strand They are then thoroughly washed to get rid of any unbounded sequences.
5. Collection and analysis
- The data collection is carried out using microarray scanners.
- The scanner consists of a laser computer as well as camera. The laser stimulated fluorescence in the cDNA creating signals.
- When the laser scans a range, the camera takes the images that are produced.
- The computer then will store the data and provide the results right away. The results can then be analyzed.
- The intensities of the colors for each spot determines what is the characteristic of the gene present in that specific spot.
Factors to consider in designing DNA Microarray experiments
When designing DNA microarray experiments, several factors need to be considered to ensure reliable and meaningful results. Here are some important factors to keep in mind:
- Controlled Experiments: It is crucial to conduct a sufficient number of controlled experiments to validate the methodology and ensure the reliability of the results. By including appropriate controls, such as negative controls or known positive controls, you can assess the specificity and accuracy of the microarray experiment.
- Duplicate Spotting and Replicate Chips: For custom-designed spotted microarrays, it is recommended to perform duplicate spotting of each probe and include replicate chips. This helps in assessing the reproducibility of the results and minimizing any potential technical variability.
- Pilot Studies: Before conducting large-scale “mega chip” experiments, it is advisable to perform pilot studies. These smaller-scale experiments can help optimize the experimental conditions, validate the experimental design, and identify any potential challenges or issues that may arise.
- Replication: Designing experiments without replication can limit the interpretability of the results. It is essential to include replicates to account for biological and technical variability. Without replication, it is difficult to draw meaningful conclusions or make reliable inferences from a single unsuccessful experiment.
- Simple Experimental Designs: Designing simple experiments with one or two factors (e.g., treatment vs. untreated) can enhance the interpretability of the results. Complex designs with multiple factors can increase the complexity of the analysis and may require more advanced statistical methods.
- Recognition of Measurement Mistakes: It is important to be vigilant and recognize potential measurement mistakes. This includes identifying technical issues, such as poor spot quality, hybridization artifacts, or labeling errors. Regular quality control checks and attention to detail during the experimental process can help identify and mitigate measurement mistakes.
- Quality of Data in Databases: If utilizing existing databases for analysis or comparison, ensure that the quality of the data is guaranteed. It is essential to assess the reliability and validity of the data sources to avoid potential biases or inaccuracies in the analysis.
- Collaboration with Statisticians: Collaborating with statisticians or colleagues experienced in statistical analysis is highly beneficial during the design stages of your research. They can provide valuable insights into appropriate experimental designs, sample size determination, statistical tests, and data analysis approaches, ensuring robust and sound statistical analysis of the microarray data.
Applications of DNA Microarray
DNA microarrays have a wide range of applications across various fields. Here are some key applications of DNA microarrays:
- Transcriptomics and Proteomics: DNA microarrays can be used to study the transcriptomes and proteomes of patients, providing insights into gene expression patterns and protein profiles associated with various diseases or conditions.
- Pathogenic and Genetic Illnesses: DNA microarrays are valuable tools for determining the presence of pathogenic microorganisms and genetic illnesses in humans. By using specific probes, microarrays can identify the microbial species or detect genetic variations associated with diseases.
- Environmental Microbiology: Microarrays with species-specific probes enable the identification and characterization of microbial communities present in the environment. This application helps in understanding microbial diversity, ecological roles, and potential implications for environmental health.
- Single Nucleotide Polymorphism (SNP) Analysis: DNA microarrays facilitate SNP analysis, allowing researchers to identify genetic variations and study their associations with diseases, population genetics, or drug responses.
- Gene Expression Profiling: DNA microarrays enable the determination of gene expression levels in specific cells or tissues at different time points. This application aids in understanding gene regulation, biological processes, and disease mechanisms.
- DNA Mutations and Genomic Alterations: Microarrays can be used to study DNA mutations, such as copy number variations, genomic gains, and losses. This information helps in characterizing genetic disorders, cancer genomics, and identifying potential therapeutic targets.
- Clinical Diagnostics: DNA microarrays have immense potential for diagnosing diseases. By analyzing gene expression profiles or detecting specific genetic variations, microarrays can aid in disease diagnosis, prognosis, and personalized medicine approaches.
- Drug Discovery: Microarrays play a crucial role in drug discovery by evaluating the effects of candidate drugs on gene expression profiles or identifying potential drug targets. This application accelerates the identification and development of new therapeutic agents.
- Functional Genomics and Proteomics: Microarrays facilitate the study of functional genomics and proteomics by providing insights into gene functions, protein interactions, and cellular pathways. They aid in deciphering biological processes and regulatory networks.
- Toxicological Research: DNA microarrays are used in toxicogenomics to assess the effects of toxins or drugs on gene expression profiles. This application helps in understanding the molecular mechanisms of toxicity and identifying biomarkers for toxicological evaluation.
- Pharmacogenomics and Theranostics: Microarrays contribute to pharmacogenomic research, which focuses on understanding how genetic variations influence drug responses. Additionally, microarrays enable the development of theranostics, combining diagnostics and therapeutics tailored to individual patients.
Advantages of DNA Microarray
DNA microarrays offer several advantages that contribute to their widespread use in genomic research and diagnostics:
- High-Throughput Analysis: DNA microarrays enable the simultaneous analysis of thousands of genes or sequences in a single experiment. This high-throughput capability allows researchers to obtain a comprehensive view of gene expression patterns or genomic variations across the genome quickly and efficiently.
- Real-Time Data: By providing data for thousands of genes in real time, DNA microarrays offer insights into the dynamic nature of gene expression. Researchers can monitor changes in gene expression levels under different experimental conditions or over time, providing valuable information about cellular processes and responses to stimuli.
- Multiplexed Results: A single DNA microarray experiment can generate a wealth of data, allowing researchers to study multiple genes or genomic regions simultaneously. This multiplexing capability saves time and resources by reducing the number of individual experiments required to obtain comprehensive genomic information.
- Rapid Results: DNA microarrays provide a fast and efficient method for obtaining results. Once the samples are hybridized to the microarray, the analysis can be performed relatively quickly, allowing researchers to obtain gene expression profiles or genomic information in a timely manner.
- Disease and Cancer Research: DNA microarrays hold great promise in the discovery of cures for diseases, including cancer. By analyzing gene expression patterns in diseased tissues or cells, researchers can identify potential therapeutic targets, biomarkers for diagnosis or prognosis, and gain insights into the underlying molecular mechanisms of diseases.
- Gene Expression Profiling: DNA microarrays are widely used to study gene expression across different tissues, developmental stages, or experimental conditions. They provide a valuable tool for identifying genes that are upregulated or downregulated in response to specific stimuli, helping researchers understand the functional roles of genes and their involvement in biological processes.
- Study of Different DNA Regions: DNA microarrays can be designed to study various regions of DNA, such as promoters, enhancers, or specific gene sequences. This flexibility allows researchers to investigate different aspects of gene expression regulation, DNA methylation patterns, or genetic variations within a population.
Disadvantages of DNA Microarray
DNA microarrays have several disadvantages that should be considered:
- Data Processing Time: The volume of information generated from each DNA microarray is substantial, resulting in a significant amount of time required for data processing and analysis. The large-scale nature of the data poses challenges in handling and interpreting the results.
- Complexity of Results: Interpreting the results of DNA microarray experiments can be challenging. The output is often presented as complex patterns or images that require expertise and specialized software for analysis. Additionally, the results obtained are not always quantitative, making it difficult to obtain precise measurements.
- Lack of Reproducibility: Reproducibility can be a concern in DNA microarray experiments. Factors such as sample preparation, labeling techniques, hybridization conditions, and data analysis methods can introduce variability, leading to inconsistent results when the experiments are repeated.
- High Cost: DNA microarray technology can be expensive, particularly when using commercial arrays or custom-designed arrays. The cost includes the production of the arrays, the required equipment and infrastructure, and the expertise needed to perform and analyze the experiments. This cost can be a limiting factor for many researchers.
- Indirect Measurement: DNA microarrays provide an indirect measure of the relative concentration of nucleic acid sequences. The hybridization signals obtained reflect the binding affinity between the probe and the target sequences rather than direct quantification. This indirect measurement can introduce variability and imprecision in the results.
- Design Challenges: Designing DNA microarrays, especially for complex genomes like mammals, can be challenging. Ensuring that multiple DNA or RNA sequences that are related do not connect to the same probe is difficult, which can impact the accuracy and specificity of the results.
- Specificity Limitations: DNA microarrays are designed to detect only the sequences they were specifically designed for. They may not capture all possible variations or mutations in the target DNA or RNA. This limited coverage can lead to potential gaps in the analysis and may miss important information.
- Shelf Life: DNA chips have a limited shelf life due to the degradation of the probes over time. The reagents and materials used in the arrays can degrade, affecting the performance and reliability of the microarrays. This limits the storage and usability of pre-manufactured arrays.
- Complexity of Analysis: The production of a large number of results in a single DNA microarray experiment requires extensive and complex data analysis. Analyzing the data, identifying significant patterns, and extracting meaningful information can be time-consuming and require specialized bioinformatics skills.
FAQ
What colors are used in a dna microarray?
Based on how the DNA binds together, each spot will appear red, green, or yellow (a combination of red and green) when scanned with a laser.
what do dna microarray assays allow scientists to study regarding gene expression?
These assays allow scientists to identify networks of gene expression across an entire genome.
in a genome-wide expression study using a dna microarray assay, what is each well used to detect?
DNA microarrays are being used to detect single nucleotide polymorphisms (SNPs) of our genome (Hap Map project), aberrations in methylation patterns, alterations in gene copy-number, alternative RNA splicing, and pathogen detection.
References
- Molecular Biotechnology – Second Edition, S. B. Primrose, Blackwell Science Inc., ASIN: 0632030534
- Introduction to Biotechnology, Brown, C.M., Campbell, I and Priest, F.G. Panima Publishing Corporation, 2005. ISBN : 81-86535-42-X
- DNA and Biotechnology, Fitzgerald-Hayes, M. And Reichsman, F. 2nd Amsterdam : Elsevier, 2010. ISBN : 0-12-048930-5
- Molecular Biotechnology : Principles and Applications of Recombinant DNA, Glick, B.R., Pasternack, Jack, J and Patten, Cheryl, L (Eds)., 4th Washington., ASM Press, 2010. ISBN : 1-55581-498-4
- Molecular Biology and Biotechnology : a guide for students, Krauzer, H. And Massey, A.(Eds) 3rd Washington DC : ASM Press, 2008, ISBN : 978-155581-4724
- Recombinant DNA and Biotechnology : a guide for teachers, Kreuzer, H and Massey, A. 2nd Washington : ASM Press, 2001, ISBN : 155581-175-2
- Khelurkar, Vaibhav & Ingle, Krishnananda & Padole, Dipika. (2017). DNA Microarray: Basic Principle and It’s Applications. 488-490.
- Anandhavalli, M. & Mishra, Chandan & Ghose, Mrinal. (2009). Analysis of Microarray Image Spots Intensity: A Comparative Study. International Journal of Computer Theory and Engineering. 1. 1793-8201. 10.7763/IJCTE.2009.V1.104.
- Bumgarner R. Overview of DNA microarrays: types, applications, and their future. Curr Protoc Mol Biol. 2013 Jan;Chapter 22:Unit 22.1.. doi: 10.1002/0471142727.mb2201s101. PMID: 23288464; PMCID: PMC4011503.
- Trevino, V., Falciani, F. & Barrera-Saldaña, H.A. DNA Microarrays: a Powerful Genomic Tool for Biomedical and Clinical Research. Mol Med 13, 527–541 (2007). https://doi.org/10.2119/2006-00107.Trevino
- Annual Review of Biomedical Engineering Vol. 4:129-153 (Volume publication date August 2002) https://doi.org/10.1146/annurev.bioeng.4.020702.153438
- https://learn.genetics.utah.edu/content/labs/microarray/
- https://www.sciencedirect.com/topics/medicine-and-dentistry/dna-microarray
- http://www.premierbiosoft.com/tech_notes/microarray.html
- https://bitesizebio.com/7206/introduction-to-dna-microarrays/
- https://courses.cs.washington.edu/courses/cse527/04au/slides/lect02.pdf
- https://ib.bioninja.com.au/options/untitled/b4-medicine/dna-microarrays.html
- https://www.omicsonline.org/open-access/dna-microarray-applications-and-limitations-118333.html
- https://www.biologyexams4u.com/2014/05/dna-microarray-steps-procedure.html
- https://www.thesciencenotes.com/dna-microarray-principle-types-procedure-and-applications/
- https://www.slideshare.net/Haddies/dna-microarray-dna-chips
- https://www.onlinebiologynotes.com/dna-microarray-principle-types-and-steps-involved-in-cdna-microarrays/
- https://www.slideshare.net/SanjaySinhmar/dna-microarray-31210546
- https://bio.as.uky.edu/sites/default/files/Overview%20of%20DNA%20microarrays.pdf
- https://www.tandfonline.com/doi/pdf/10.1080/10245330701283967
- https://www.biotechnologynotes.com/microarrays/microarray-meaning-production-and-types-biotechnology/597
- http://revistabionatura.com/files/2017.02.03.13.pdf
- https://www.csus.edu/indiv/r/rogersa/bio181/microarrays.pdf
- http://ecoursesonline.iasri.res.in/mod/page/view.php?id=5073
- https://www.genome.gov/about-genomics/fact-sheets/DNA-Microarray-Technology#:~:text=in%20BRCA1%20alone.-,The%20DNA%20microarray%20is%20a%20tool%20used%20to%20determine%20whether,used%20to%20make%20computer%20microchips.
- https://en.wikipedia.org/wiki/DNA_microarray
- https://www.news-medical.net/life-sciences/DNA-microarray.aspx
- https://www.news-medical.net/life-sciences/Types-of-Microarray.aspx#:~:text=Based%20on%20the%20types%20of%20probes%20used%2C%20microarrays%20are%20of,part%20of%20its%20detection%20system.
