Table of Contents
Before going through Eukaryotic Translation steps, please take a look at our previous article Prokaryotic Translation Steps, Requirements, to check the components required for the translation procedure.
Protein synthesis in eukaryotes, which includes organisms such as animals, plants, fungi, and protists, involves a complex process that occurs in two main stages: transcription and translation. Here’s an overview of protein synthesis in eukaryotes:
- Transcription:
- The process begins in the nucleus, where DNA is located. The DNA sequence of a specific gene that encodes a protein serves as a template.
- RNA polymerase, an enzyme, binds to the DNA at the gene’s promoter region and initiates transcription.
- RNA polymerase moves along the DNA strand, unwinding it and synthesizing a complementary RNA molecule called pre-mRNA.
- As RNA polymerase progresses, it reads the DNA sequence and incorporates complementary RNA nucleotides (adenine [A], cytosine [C], guanine [G], and uracil [U]) into the growing pre-mRNA molecule.
- After reaching the terminator region, RNA polymerase completes transcription, and the pre-mRNA molecule is formed.
- RNA Processing:
- In eukaryotes, pre-mRNA undergoes several modifications before it can serve as a functional messenger RNA (mRNA) molecule.
- These modifications include the removal of non-coding regions called introns through a process called splicing. The remaining coding regions, called exons, are joined together.
- A modified guanine nucleotide, called a 5′ cap, is added to the 5′ end of the mRNA, providing stability and aiding in translation initiation.
- A poly-A tail, a string of adenine nucleotides, is added to the 3′ end of the mRNA, which helps protect it from degradation.
- Translation:
- Translation occurs in the cytoplasm, specifically on ribosomes. Ribosomes consist of large and small subunits, and they provide the platform for protein synthesis.
- The mRNA, carrying the genetic information, binds to a ribosome, which scans the mRNA for a start codon (usually AUG) that indicates the beginning of the protein-coding sequence.
- Transfer RNA (tRNA) molecules, each carrying a specific amino acid, recognize the codons on the mRNA through complementary base pairing.
- As the ribosome moves along the mRNA, tRNA molecules bring in the corresponding amino acids and form peptide bonds between them, creating a growing polypeptide chain.
- The ribosome continues elongating the polypeptide chain until it reaches a stop codon (UAA, UAG, or UGA), which signals the end of translation.
- The newly synthesized polypeptide chain is released, and the ribosome, mRNA, and tRNA dissociate.
After translation, the polypeptide chain may undergo additional modifications, such as folding, cleavage, or chemical modifications, to form the functional protein. The protein is then transported to its target location within the cell or secreted outside of the cell to carry out its specific function.
Overall, protein synthesis in eukaryotes involves the coordinated action of DNA, RNA, and ribosomes to convert the genetic information encoded in DNA into functional proteins.
Eukaryotic ribosomes
- Eukaryotic ribosomes are complex cellular structures responsible for protein synthesis in eukaryotic cells. They are composed of ribosomal RNA (rRNA) molecules and various proteins. Eukaryotic ribosomes are larger and more structurally complex compared to their prokaryotic counterparts.
- The eukaryotic ribosome is made up of two subunits: the large subunit (60S) and the small subunit (40S). The numbers represent the sedimentation coefficient, which is a measure of the rate at which the ribosomes sediment during centrifugation.
- The large subunit contains three rRNA molecules (28S, 5.8S, and 5S) and multiple proteins, while the small subunit contains a single rRNA molecule (18S) and additional proteins. These rRNA molecules play crucial roles in the catalytic activity and structural integrity of the ribosome.
- Eukaryotic ribosomes are primarily located in the cytoplasm, where they carry out the translation of mRNA into proteins. However, some ribosomes also associate with the rough endoplasmic reticulum (ER), forming the rough ER, which is involved in protein synthesis and modification for secretion or membrane integration.
- During translation, the small subunit of the ribosome binds to the mRNA, and the large subunit joins to form a functional ribosome. Transfer RNA (tRNA) molecules, carrying specific amino acids, recognize and bind to the codons on the mRNA through complementary base pairing. The ribosome catalyzes the formation of peptide bonds between the amino acids, creating a growing polypeptide chain.
- Eukaryotic ribosomes are dynamic and can undergo various modifications and interactions with other proteins and factors to regulate translation and ensure accurate protein synthesis. These processes play essential roles in cell growth, development, and maintaining cellular homeostasis.
Ribosomal Sites for Protein Synthesis
During protein synthesis in ribosomes, there are three key sites involved: the A site (aminoacyl site), the P site (peptidyl site), and the E site (exit site). These sites are where the ribosome interacts with different molecules and perform specific functions in the process of translation.
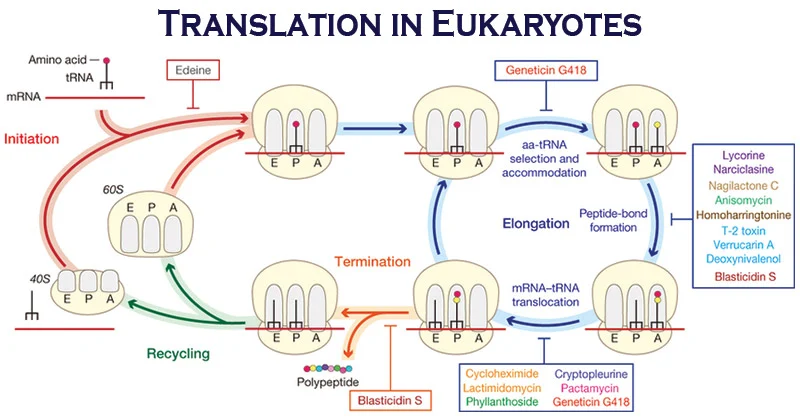
- A site (aminoacyl site): The A site is the binding site for incoming aminoacyl-tRNA (charged tRNA). It is where the tRNA carrying the next amino acid to be added to the growing polypeptide chain enters the ribosome. The codon-anticodon recognition occurs in the A site. Once the correct aminoacyl-tRNA is bound, peptide bond formation can take place.
- P site (peptidyl site): The P site is where the tRNA carrying the growing polypeptide chain is bound. It holds the peptidyl-tRNA, which is the tRNA linked to the polypeptide chain. The P site is also the site of peptide bond formation between the amino acid on the peptidyl-tRNA and the amino acid on the aminoacyl-tRNA in the A site.
- E site (exit site): The E site is where the now uncharged tRNA, which has donated its amino acid to the growing polypeptide chain, exits the ribosome. The E site is involved in the release of the tRNA from the ribosome after its role in protein synthesis is completed. The ribosome then moves along the mRNA, shifting the remaining tRNAs from A to P and E to A sites, respectively.
The movement of tRNAs through these ribosomal sites—A, P, and E—is facilitated by ribosomal translocation, where the ribosome advances along the mRNA in a processive manner.
The coordinated interplay of these ribosomal sites is crucial for accurate protein synthesis, ensuring that the correct amino acids are incorporated into the growing polypeptide chain based on the mRNA template.
Differences between eukaryotic and prokaryotic Protein Synthesis
- Ribosome Sedimentation Coefficient: The sedimentation coefficient of a prokaryotic ribosome is 70S, with subunits of 30S (small) and 50S (large). In contrast, eukaryotic ribosomes have a sedimentation coefficient of 80S, with subunits of 40S (small) and 60S (large).
- Ribosomal Subunit Composition: Eukaryotic ribosomal subunits are more complex in composition compared to prokaryotic subunits. However, the basic functions of each subunit remain similar between eukaryotes and prokaryotes.
- Monocistronic vs. Polycistronic mRNA: In eukaryotes, each mRNA molecule is usually monocistronic, meaning it encodes a single protein. Prokaryotes often have polycistronic mRNA, where a single mRNA can encode multiple proteins with separate initiation and termination codons for each coding sequence.
- Initiation Factors: Eukaryotes require a more extensive set of initiation factors compared to prokaryotes. At least nine distinct eukaryotic initiation factors (eIFs) are involved in the initiation of protein synthesis, while prokaryotes use three initiation factors (IFs).
- Initiating Amino Acid: In eukaryotes, the initiating amino acid for protein synthesis is methionine, whereas prokaryotes use N-formylmethionine (fMet) as the initiating amino acid. Eukaryotes have a specific initiator tRNA called Met-tRNAimet for initiation, distinct from the tRNA recognizing methionine at internal positions in the mRNA.
- Initiation Process: Prokaryotic initiation involves a Shine-Dalgarno sequence located upstream of the AUG initiation codon, serving as the binding site for the 30S ribosomal subunit. In contrast, most eukaryotic mRNAs lack Shine-Dalgarno sequences. Instead, a 40S ribosomal subunit attaches to the mRNA’s 5′ end and scans downstream (5′ to 3′ direction) until it finds the AUG initiation codon. This process is known as scanning.
- Helicase Requirement: Prokaryotic translation does not require helicase since protein synthesis can begin while mRNA is still being synthesized. In eukaryotes, transcription in the nucleus and translation in the cytoplasm are separate events, allowing time for mRNA secondary structure to form. Thus, helicase is involved in unwinding the secondary structures during eukaryotic translation.
- Compartmentalization: In eukaryotes, protein synthesis occurs in the cytoplasm, where ribosomes are distributed, as well as on the rough endoplasmic reticulum (ER) for proteins destined for secretion or membrane integration. In contrast, prokaryotes lack membrane-bound organelles, and protein synthesis takes place in the cytoplasm.
- Ribosome Size: Eukaryotic ribosomes are larger than prokaryotic ribosomes. Eukaryotes have 80S ribosomes, consisting of a 60S large subunit and a 40S small subunit. Prokaryotes have 70S ribosomes, composed of a 50S large subunit and a 30S small subunit. The S unit represents the sedimentation coefficient during centrifugation.
- RNA Polymerases: Eukaryotes have multiple RNA polymerases responsible for transcription of different types of RNA, including mRNA, rRNA, and tRNA. Prokaryotes have a single RNA polymerase that synthesizes all types of RNA.
- Transcription-Coupled Translation: In prokaryotes, transcription and translation occur simultaneously since there is no nuclear membrane separating the two processes. This allows for rapid protein synthesis. In eukaryotes, transcription occurs in the nucleus, and the pre-mRNA is processed and transported to the cytoplasm before translation can occur. Thus, transcription and translation are spatially separated in eukaryotes.
- mRNA Processing: Eukaryotic mRNA undergoes extensive processing, including splicing, 5′ capping, and 3′ polyadenylation, before it can be used for translation. Prokaryotic mRNA lacks these modifications.
- Shine-Dalgarno Sequence: Prokaryotic mRNA contains a Shine-Dalgarno sequence, a specific ribosome-binding site located upstream of the start codon. This sequence helps position the ribosome correctly during translation initiation. Eukaryotic mRNA, on the other hand, typically contains a 5′ cap and an mRNA sequence called the Kozak sequence near the start codon to aid in translation initiation.
Eukaryotic Translation
- Eukaryotic translation is the biological process by which messenger RNA is translated into proteins in eukaryotes.
- It consists of four phases: gene regulation, elongation, termination, and recycling.
Eukaryotic initiation factor
Eukaryotic initiation factors (eIFs) are proteins or protein complexes involved in the initiation phase of eukaryotic translation. These proteins help stabilize the formation of ribosomal preinitiation complexes around the start codon and are an important input for post-transcription gene regulation.
| Factor | Function |
| eIF1 and eIF1A | eIF1 and eIF1A both bind to the 40S ribosome subunit-mRNA complex. Together they induce an “open” conformation of the mRNA binding channel, which is crucial for scanning, tRNA delivery, and start codon recognition. |
| eIF2 | eIF2 is the main protein complex responsible for delivering the initiator tRNA to the P-site of the preinitiation complex, as a ternary complex containing Met-tRNAiMet and GTP (the eIF2-TC). |
| eIF3 | eIF3 independently binds the 40S ribosomal subunit, multiple initiation factors, and cellular and viral mRNA. |
| eIF4 | The eIF4F complex is composed of three subunits: eIF4A, eIF4E, and eIF4G. Each subunit has multiple human isoforms and there exist additional eIF4 proteins: eIF4B and eIF4H. eIF4G is a 175.5-kDa scaffolding protein that interacts with eIF3 and the Poly(A)-binding protein (PABP), as well as the other members of the eIF4F complex. eIF4E recognizes and binds to the 5′ cap structure of mRNA, while eIF4G binds PABP, which binds the poly(A) tail, potentially circularizing and activating the bound mRNA. eIF4A – a DEAD box RNA helicase – is important for resolving mRNA secondary structures. |
| eIF5 | eIF5 is a GTPase-activating protein, which helps the large ribosomal subunit associate with the small subunit. It is required for GTP-hydrolysis by eIF2 and contains the unusual amino acid hypusine. |
| eIF5A | eIF5A is the eukaryotic homolog of EF-P. It helps with elongation and also plays a role in termination. |
| eIF5B | eIF5B is a GTPase, and is involved in assembly of the full ribosome. It is the functional eukaryotic analog of bacterial IF2. |
| eIF6 | eIF6 performs the same inhibition of ribosome assembly as eIF3, but binds with the large subunit. |
Elongation factor for Eukaryotic Translation
Elongation factors are a set of proteins that function at the ribosome, during protein synthesis, to facilitate translational elongation from the formation of the first to the last peptide bond of a growing polypeptide.
| Bacterial | Eukaryotic/Archaeal | Function |
|---|---|---|
| EF-Tu | eEF-1 subunit α | mediates the entry of the aminoacyl tRNA into a free site of the ribosome. |
| EF-Ts | eEF-1 subunit βγ | serves as the guanine nucleotide exchange factor for EF-Tu, catalyzing the release of GDP from EF-Tu. |
| EF-G | eEF-2 | catalyzes the translocation of the tRNA and mRNA down the ribosome at the end of each round of polypeptide elongation. Causes large conformation changes. |
| EF-P | EIF5A | possibly stimulates formation of peptide bonds and resolves stalls. |
Steps of Eukaryotic Translation
The eukaryotic translation is completed in the following steps;
- Initiation of Eukaryotic Translation
- Elongation of peptide chain
- Termination of peptide chain
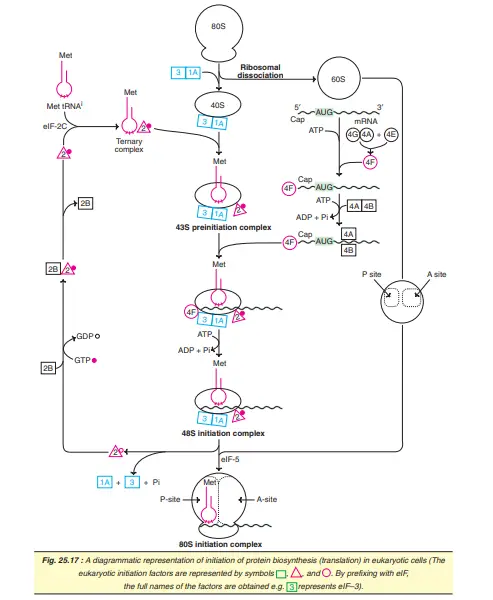
Initiation of Eukaryotic Translation
The initiation of translation in eukaryotes is complex, involving at least ten eukaryotic initiation factors (eIFs). Some of the eIFs contain multiple (3-8) subunits. The process of translation initiation can be divided into four steps.
- Ribosomal dissociation.
- Formation of 43S preinitiation complex.
- Formation of 48S initiation complex.
- Formation of 80S initiation complex.
Ribosomal dissociation
- The 80S ribosome dissociates to form 40S and 60S subunits.
- Two initiating factors namely eIF3 and eIF-1A bind to the newly formed 40S subunit, and thereby block its reassociation with 60S subunit. For this reason, some workers name eIF-3 as anti-association factor.
Formation of 43S preinitiation complex
- A ternary complex containing met-tRNAi and eIF-2 bound to GTP attaches to 40S ribosomal subunit to form 43S preinitiation complex.
- The presence of eIF-3 and eIF-1A stabilizes this complex (Note : Met-tRNA is specifically involved in binding to the initiation condon AUGs; hence the superscripti is used in mettRNAi).
Formation of 48S initiation complex
- The binding of mRNA to 43S preinitiation complex results in the formation of 48S initiation complex through the intermediate 43S initiation complex. This, however, involves certain interactions between some of the eIFs and activation of mRNA.
- eIF-4F complex is formed by the association of eIF-4G, eIF-4A with eIF-4E.
- The so formed eIF-4F (referred to as cap binding protein) binds to the cap of mRNA.
- Then elF-4A and elF-4B bind to mRNA and reduce its complex structure.
- This mRNA is then transferred to 43S complex.
- For the appropriate association of 43S preinitiation complex with mRNA, energy has to be supplied by ATP.
Recognition of initiation codon:
- The ribosomal initiation complex scans the mRNA for the identification of appropriate initiation codon. 5c-AUG is the initiation codon and its recognition is facilitated by a specific sequence of nucleotides surrounding it.
- This marker sequence for the identification of AUG is called as Kozak consensus sequence.
- In case of prokaryotes the recognition sequence of initiation codon is referred to as Shine- Dalgarno sequence.
Formation of 80S initiation complex
- 48S initiation complex binds to 60S ribosomal subunit to form 80S initiation complex.
- The binding involves the hydrolysis of GTP (bound to eIF-2). This step is facilitated by the involvement of eIF-5.
- As the 80S complex is formed, the initiation factors bound to 48S initiation complex are released and recycled.
- The activation of eIF-2 requires eIF-2B (also called as guanine nucleotide exchange factor) and GTP.
- The activated eIF-2 (i.e. bound to GTP) requires eIF2C to form the ternary complex.
Regulation of initiation
- The eIF-4F, a complex formed by the assembly of three initiation factors controls initiation, and thus the translation process.
- eIF4E, a component of eIF-4F is primarily responsible for the recognition of mRNA cap. And this step is the rate-limiting in translation.
- eIF-2 which is involved in the formation of 43S preinitiation complex also controls protein biosynthesis to some extent.
Elongation of Eukaryotic Translation
Ribosomes elongate the polypeptide chain by a sequential addition of amino acids. The amino acid sequence is determined by the order of the codons in the specific mRNA. Elongation, a cyclic process involving certain elongation factors (EFs), may be divided into three steps.
- Binding of aminoacyl t-RNA to A-site.
- Peptide bond formation.
- Translocation.
Binding of aminoacyl—tRNA to A-site
- The 80S initiation complex contains met-tRNAi in the P-site, and the A-site is free.
- Another aminoacyl-tRNA is placed in the A-site. This requires proper codon recognition on the mRNA and the involvement of elongation factor 1a (EF-Ia) and supply of energy by GTP.
- As the aminoacyl-tRNA is placed in the A-site, EF-1D and GDP are recycled to bring another aminoacyl-tRNA.
Peptide bond formation
- The enzyme peptidyltransferase catalyses the formation of peptide bond.
- The activity of this enzyme lies on 28S RNA of 60S ribosomal subunit. It is therefore the rRNA (and not protein) referred to as ribozyme that catalyses the peptide bond formation.
- As the amino acid in the aminoacyl-tRNA is already activated, no additional energy is required for peptide bond formation. The net result of peptide bond formation is the attachment of the growing peptide chain to the tRNA in the A-site.
Translocation
- As the peptide bond formation occurs, the ribosome moves to the next codon of the mRNA (towards 3c-end). This process called translocation, basically involves the movement of growing peptide chain from A-site to P-site.
- Translocation requires EF-2 and GTP.
- GTP gets hydrolysed and supplies energy to move mRNA. EF-2 and GTP complex recycles for translocation.
- In recent years, another site namely exit site (E-site) has been identified in eukaryotes. The deacylated tRNA moves into the E-site, from where it leaves the ribosome.
- In case of prokaryotes, the elongation factors are different, and they are EF-Tu, EF-Ts (in place of of EF-1a) and EF-G (instead of EF-2).
Incorporation of amino acids
- It is estimated that about six amino acids per second are incorporated during the course of elongation of translation in eukaryotes.
- In case of prokaryotes, as many as 20 amino acids can be incorporated per second.
- Thus the process of protein/polypeptide synthesis in translation occurs with great speed and accuracy.
Termination of Eukaryotic Translation
Termination is a simple process when compared to initiation and elongation. After several cycles of elongation, incorporating amino acids and the formation of the specific protein/ polypeptide molecule, one of the stop or termination signals (UAA, UAG and UCA) terminates the growing polypeptide.
The termination codons which act as stop signals do not have specific tRNAs to bind. As theb termination codon occupies the ribosomal A-site, the release factor namely eRF recognizes the stop signal. eRF-GTP complex, in association with the enzyme peptidyltransferase, cleaves the peptide bond between the polypeptide and the tRNA occupying P-site. In this reaction, a water molecule, instead of an amino acid is added. This hydrolysis releases the protein and tRNA from the P-site. The 80S ribosome dissociates to form 40S and 60S subunits which are recycled. The mRNA is also released.
Where does translation occur in eukaryotes?/where does translation take place in eukaryotic cells?
Translation in eukaryotes occurs in the cytoplasm. After transcription of the DNA into mRNA is completed in the nucleus, the mRNA is processed and transported out of the nucleus into the cytoplasm. In the cytoplasm, ribosomes, which are the cellular machinery responsible for protein synthesis, bind to the mRNA and initiate the process of translation. The ribosomes can be found freely in the cytoplasm or attached to the rough endoplasmic reticulum (ER) in the case of proteins destined for secretion or membrane integration. Therefore, translation primarily takes place in the cytoplasm of eukaryotic cells.
Eukaryotic vs Prokaryotic translation
| Aspect | Eukaryotic Translation | Prokaryotic Translation |
|---|---|---|
| Ribosome Size | 80S (composed of 40S small subunit and 60S large subunit) | 70S (composed of 30S small subunit and 50S large subunit) |
| mRNA Processing | Extensive processing, including splicing, capping, and polyadenylation | Minimal processing, lacking splicing and extensive modifications |
| mRNA Structure | Monocistronic (encodes a single protein) | Polycistronic (encodes multiple proteins) |
| Initiation Factors | Require at least nine distinct eukaryotic initiation factors (eIFs) | Require three initiation factors (IFs) |
| Initiating Amino Acid | Methionine | N-formylmethionine (fMet) |
| Initiator tRNA | Met-tRNAimet | fMet-tRNAfMet |
| Ribosome Binding Site | Ribosome attaches to the 5′ cap of mRNA and scans to find the start codon | Shine-Dalgarno sequence upstream of the start codon |
| Ribosome Localization | Cytoplasm and rough endoplasmic reticulum (ER) | Cytoplasm |
| mRNA Secondary Structure | Helicase may be required for unwinding secondary structures during translation | Translation can begin while mRNA is still being synthesized |
FAQ
What is eukaryotic translation?
Eukaryotic translation is the process by which proteins are synthesized in eukaryotic cells using the genetic information encoded in mRNA molecules.
Where does eukaryotic translation occur?
Eukaryotic translation occurs in the cytoplasm, where ribosomes are distributed, and on the rough endoplasmic reticulum (ER) for proteins destined for secretion or membrane integration.
What is the role of ribosomes in eukaryotic translation?
Ribosomes are responsible for protein synthesis in eukaryotic cells. They facilitate the assembly of amino acids into polypeptide chains based on the instructions provided by mRNA.
What are the subunits of eukaryotic ribosomes?
Eukaryotic ribosomes are composed of a large subunit (60S) and a small subunit (40S), which come together during translation to form a functional ribosome.
What is the function of the small subunit in eukaryotic translation?
The small subunit of eukaryotic ribosomes binds to the mRNA and initiates the scanning process to locate the start codon.
What is the function of the large subunit in eukaryotic translation?
The large subunit of eukaryotic ribosomes catalyzes the formation of peptide bonds between amino acids, leading to the elongation of the polypeptide chain.
What are initiation factors in eukaryotic translation?
Initiation factors, known as eIFs (eukaryotic initiation factors), are proteins required for the initiation of translation in eukaryotes. They are involved in assembling the ribosome at the start codon and recruiting the initiator tRNA.
How is the genetic code deciphered during eukaryotic translation?
The genetic code is deciphered during eukaryotic translation through the complementary base pairing between the codons on mRNA and the anticodons on tRNA molecules carrying amino acids.
What is the significance of the Kozak sequence in eukaryotic translation?
The Kozak sequence is a specific sequence surrounding the start codon in eukaryotic mRNA. It helps to facilitate the recognition and accurate initiation of translation by the ribosome.
What happens during the termination phase of eukaryotic translation?
During termination, a release factor binds to the stop codon in the mRNA, leading to the release of the newly synthesized polypeptide from the ribosome. The ribosome then dissociates, and the components are recycled for subsequent rounds of translation.
Where do transcription and translation occur in eukaryotic cells?
The coding regions of eukaryotic mRNA that are translated are known as open reading frames (ORFs). An open reading frame refers to a sequence of nucleotides on the mRNA that can be translated into a polypeptide chain. These regions typically start with a start codon (usually AUG) and end with a stop codon (UAA, UAG, or UGA).
Within eukaryotic mRNA, the coding regions can be interrupted by non-coding regions called introns. Introns are non-coding sequences that are transcribed from the DNA but are removed during mRNA processing through a process called splicing. The remaining exons, which are the coding regions, are joined together to form the mature mRNA.
Therefore, after the splicing process, the mature mRNA consists of uninterrupted coding regions (exons) that can be translated into proteins by the ribosomes during eukaryotic translation.
What are coding regions of eukaryotic mrna that are translated?
The coding regions of eukaryotic mRNA that are translated are called protein-coding sequences or protein-coding regions. These regions contain the genetic information necessary to produce a protein. In eukaryotes, protein-coding sequences are typically found within exons, which are the coding regions of the gene.
During mRNA processing, pre-mRNA undergoes splicing, where introns (non-coding sequences) are removed, and exons are joined together to form mature mRNA. The mature mRNA consists of uninterrupted protein-coding sequences that can be translated into proteins.
The protein-coding regions are bounded by start codons (usually AUG) at the beginning and stop codons (UAA, UAG, or UGA) at the end. The start codon serves as the initiation point for translation, and the ribosomes read the mRNA sequence in triplets (codons), translating them into the corresponding amino acids that form the protein.
In summary, the coding regions of eukaryotic mRNA that are translated are the protein-coding sequences located within the exons, which are joined together after the removal of introns through the splicing process.
Which of the events occur during eukaryotic translation initiation?
During eukaryotic translation initiation, several key events take place. Here are the main steps involved:
Binding of the Small Ribosomal Subunit: The small ribosomal subunit (40S) binds to the mRNA molecule at the 5′ untranslated region (UTR) with the help of initiation factors.
Recognition of the Start Codon: The ribosome scans the mRNA in a 5′ to 3′ direction until it encounters the start codon, which is usually AUG (encoding methionine). The start codon is recognized by the initiator tRNA, which carries the amino acid methionine.
Assembly of the Initiation Complex: Several initiation factors assist in the assembly of the initiation complex. One of the key factors is the eukaryotic initiation factor 2 (eIF2), which is responsible for delivering the initiator tRNA (charged with methionine) to the ribosome.
Joining of the Large Ribosomal Subunit: Once the start codon is recognized, the large ribosomal subunit (60S) joins the small subunit, forming a functional ribosome ready for protein synthesis.
GTP Hydrolysis: During initiation, GTP (guanosine triphosphate) is hydrolyzed to GDP (guanosine diphosphate) and inorganic phosphate (Pi) as a part of the activation process for ribosome assembly and subsequent steps.
These events collectively ensure the proper initiation of protein synthesis in eukaryotic cells. Initiation factors play crucial roles in coordinating the assembly of ribosomal subunits, positioning the start codon, and delivering the initiator tRNA to the ribosome, allowing the translation process to proceed.
References
- https://microbiologynote.com/prokaryotic-translation-steps-requirements/
- https://en.wikipedia.org/wiki/Translation

Wow valuable information. ..🙏
Wow Informative information. ..🙏