Table of Contents
What is mRNA?
This sort of RNA acts by transferring genetic material into ribosomes and transmitting instructions regarding the types of proteins that body cells require. Based on their roles, these RNA types are known as messenger RNA. Therefore, mRNA plays an essential function in the transcription process or during protein synthesis.
- Messenger ribonucleic acids (mRNAs) transmit DNA information to the protein-producing machinery of the cell.
- A three-meter-long double-stranded DNA “instruction manual” for building and maintaining the human body is tightly packed within every cell nucleus, which measures just 10 microns in diameter.
- To maintain its structure and carry out all of its duties, each cell must continuously produce cell-type-specific components (proteins).
- RNA polymerase II (RNAP II) is a multisubunit protein that reads DNA and simultaneously synthesises messenger RNA (mRNA) during the transcription process.
- mRNA molecules consist of relatively short, single strands of adenine, cytosine, guanine, and uracil bases bound together by a sugar phosphate backbone.
- When RNA polymerase finishes reading a piece of DNA, the pre-mRNA copy is processed to generate mature mRNA, which is then exported from the cell nucleus.
- In a process known as translation, ribosomes read the mRNA and translate the message into functional proteins.
- Depending on the structure and function of the newly generated protein, it will undergo further modification by the cell, be exported to the extracellular space, or remain within the cell.
- The diagram below depicts transcription (DNA->RNA) occurring in the nucleus of the cell, where the RNA polymerase II enzyme RNAP synthesises RNA.
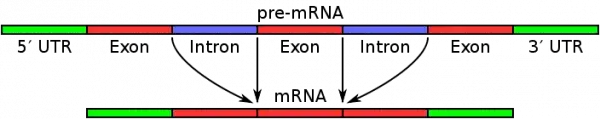
- Precursor mRNA has both introns and exons. Exons code for the amino acid sequence of proteins, while introns are deleted before to translation.
- To produce mature mRNA, the machinery of the cell eliminates “non-translatable” introns from the pre-mRNA, leaving only translatable exon regions in the mRNA.
History of mRNA
- Since the early 1950s, molecular studies showed that when proteins are made, there are molecules that are related to RNA. In one of the first reports, Jacques Monod and his team showed that RNA synthesis was needed for protein synthesis, especially when the enzyme -galactosidase was made in the bacterium E. coli.
- In 1954, Arthur Pardee also found the same kind of RNA buildup.
- Alfred Hershey, June Dixon, and Martha Chase wrote about a type of DNA that had cytosine in it, which meant it was RNA, and that disappeared quickly after being made in E. coli. This was the first record of mRNA, but it wasn’t called that at the time.
- Sydney Brenner and Francis Crick came up with the idea of mRNA on April 15, 1960, at King’s College in Cambridge, England. At the time, Francois Jacob was telling them about an experiment that Arthur Pardee, himself, and Monod had just done.
- Crick encouraged Brenner and Jacob to test this new idea right away, so they contacted Matthew Meselson at the California Institute of Technology.
- Brenner, Jacob, and Meselson did an experiment in Meselson’s lab at Caltech during the summer of 1960 that proved the existence of mRNA. In the fall of that year, Jacob and Monod came up with the name “messenger RNA” and made the first theory to explain how it works.
- In February 1961, James Watson told them that his research group was doing a similar experiment in a similar direction right behind them. Brenner and the others agreed to Watson’s request to hold off on publishing their research results until after his group’s experiment was done.
- Because of this, the articles by Brenner and Watson were published at the same time in May 1961 in the same issue of Nature. In the same month, Jacob and Monod published their theory for mRNA in the Journal of Molecular Biology.
Properties of mRNA
- In eukaryotes, the heterogeneous nuclear RNA that makes up the mRNA is made in the nucleus (hnRNA).
- When hnRNA is broken down, the functional mRNA is released. This mRNA goes into the cytoplasm to help make proteins. mRNA is made up of molecules that are big and have a short half-life.
- mRNA in eukaryotes is more stable and has a longer half-life than mRNA in prokaryotes.
- 7-methylguanosine triphosphate is added to the 5′ end of the mRNA in eukaryotes.
- It is thought that this cap stops 5′-exonucleases from breaking down mRNA by adding water.
- Also, the cap may play a role in how mRNA is recognised for protein synthesis.
- A polymer of adenylate residues (20–250 nucleotides) is found at the 3′ end of mRNA. This is called the poly (A) tail.
- This tail could make mRNA more stable and protect it from 3c-exonucleases at the same time.
- Inside the structure of mRNA molecules, you can often find modified bases like 6-methyl adenylates.
mRNA Structure
Messenger RNA, or mRNA, is a type of RNA. Ribosomes translate the single-stranded RNA template in order to make proteins.
The name “messenger RNA” was given by Jacob and Monad. It is a long molecule made up of nucleotides that are linked together.
mRNA is a long, single-stranded molecule made up of nucleotides connected by phosphodiester bonds. It has adenine, guanine, cytosine, and uracil, which are all nitrogenous bases. The RNA matches one of the DNA strands, but instead of thymine, it has uracil.

The mature mRNA is made up of the 5′ cap, the 5′ UTR, the coding region, the 3′ UTR, and the poly(A) tail.
Coding region
- It is made up of triplets of nucleotides, which are called codons.
- Each codon tells ribosomes how to make a certain amino acid, and ribosomes use codons in mRNA to make a chain of amino acids.
- The coding region starts with the start codon, which is “AUG,” and ends with one of the stop codons, which are “UAG,” “UAA,” or “UGA.” A piece of the coding sequence could also be used to control something.
Untranslated regions or UTRs
- They are in the 5′ and 3′ regions, which come before and after the coding region. There are untranslated parts before and after the start and stop codons.
- They play a part in how genes are turned on and off.
- They affect how stable RNA is, how well translation works, and where mRNA is found.
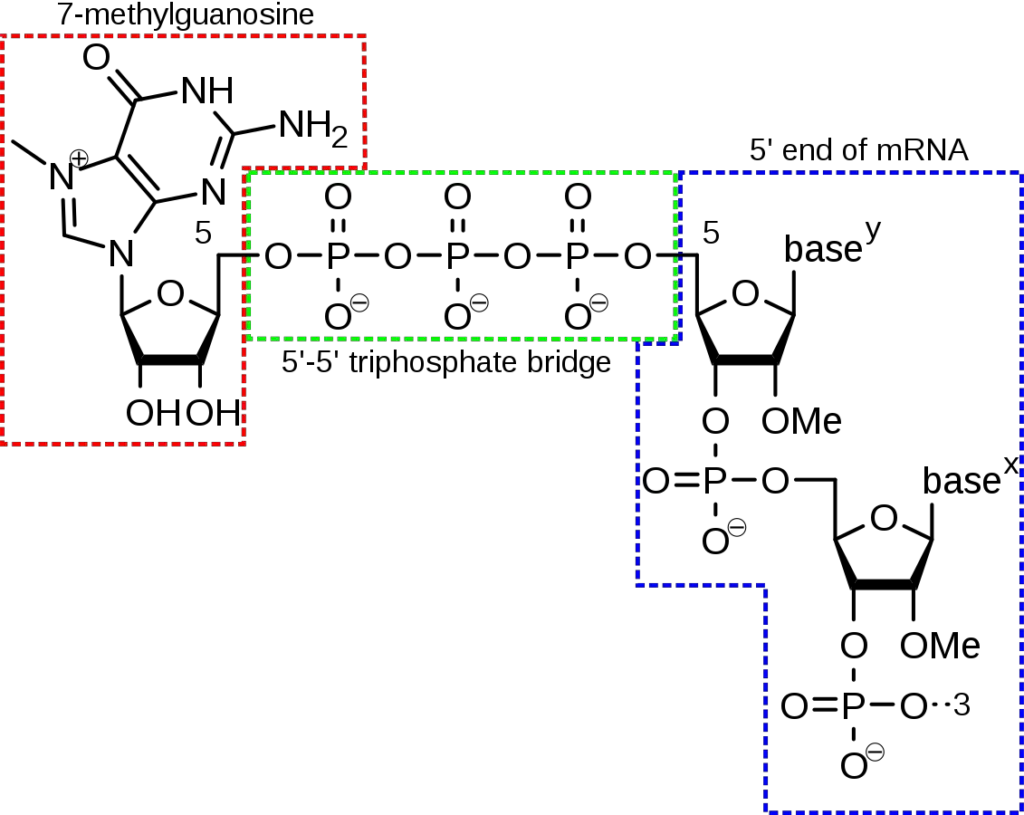
5’ Cap
- At the 5′-end, there is a cap made of methyl guanosine triphosphate.
- A 5′ cap, also called an RNA cap, an RNA 7-methylguanosine cap, or an RNA m7G cap, is a modified guanine nucleotide that has been added to the “front” or 5′ end of a eukaryotic messenger RNA soon after transcription begins.
- The 5′ cap is made up of a 7-methylguanosine residue at the end that is linked to the first nucleotide that was copied by a 5′-5′-triphosphate bond.
- It must be there for the ribosome to recognise it and for it to be safe from RNases.
- Cap addition is linked to transcription and happens at the same time as transcription. This means that each process affects the other. Shortly after the start of transcription, a cap-synthesizing complex that is part of RNA polymerase binds to the 5′ end of the mRNA that is being made.
- This complex of enzymes speeds up the chemical reactions that are needed for capping mRNA. Synthesis is a biochemical process with many steps.
Polyadenylation – Poly(A) Tail
- At the 3′ end, there is a polyadenylate tail.
- A polyadenylation is when a poly adenylyl group is chemically linked to a messenger RNA molecule.
- Most messenger RNA (mRNA) molecules in eukaryotic organisms have a polyadenylated 3′ end. However, new research has shown that short stretches of uridine (oligouridylation) are also common.
- Exonucleases can break down mRNA, but the poly(A) tail and the protein that is bound to it help protect it.
- Polyadenylation is also needed for transcription to end, for mRNA to leave the nucleus, and for translation to happen.
- mRNA can also be polyadenylated in prokaryotic organisms, where the poly(A) tails help the exonucleolytic degradation of mRNA rather than stopping it.
- Polyadenylation can happen during or right after the process of turning DNA into RNA.
- The mRNA chain is cut by an endonuclease complex that is part of RNA polymerase. This happens after transcription is done.
- After the mRNA is cut, about 250 adenosine residues are added to the free 3′ end at the site where the mRNA was cut. Polyadenylate polymerase is what makes this reaction happen.
- There can be more than one polyadenylation variant of an mRNA, just like there can be more than one way to splice an mRNA.
- Mutations can also happen at polyadenylation sites. At the poly-A addition site, the primary RNA transcript of a gene is cut, and 100–200 A’s are added to the 3′ end of the RNA.
- If this site is changed, an mRNA construct that is too long and unstable will be made.
Most eukaryotic mRNAs are monocistronic, which means that their coding region codes for only one protein. Polycistronic means that most mRNAs in prokaryotes code for more than one protein. Most of the functions of these proteins are similar, and they are controlled by a single set of promoter and operator regions. The mitochondrial genome in humans is made up of many copies.
Types of mRNA
1. Pre-mRNA and hnRNA
- Precursor mRNA, or pre-mRNA, is the primary transcript of eukaryotic mRNA as it comes off the DNA template.
- Heterogeneous nuclear RNA is the name for a group of RNAs that includes pre-mRNA (hnRNA).
- hnRNA is all the single-stranded RNA that is in the nucleus of the cell, where transcription from DNA to RNA happens. A lot of these ribonucleic acids are pre-mRNA.
- Before pre-mRNA can be turned into a protein, it needs to have some sequences taken out, or “spliced out.”
- These sequences can be taken out either by the catalytic action of the RNA itself or by a structure made of several proteins called the spliceosome.
- After this step, the pre-mRNA is considered to be an mRNA transcript that is ready to be used.
2. Monocistronic mRNA
- A monocistronic mRNA molecule is made up of the exon sequences that code for a single protein.
- It is made from a single gene (a cistron), and there is only one initiation codon and one termination codon.
- Most of the mRNAs in eukaryotes are monocistronic.
3. Bicistronic mRNA
- A molecule of mRNA that is Bicistronic has the coding sequences for two proteins in its exons.
4. Polycistronic mRNA
- Multiple proteins’ exon coding sequences are included within a polycistronic mRNA molecule.
- The majority of mRNA in bacteria and bacteriophages (viruses with bacterial hosts) is polycistronic.
- Contrary to monocistronic mRNA, this mRNA molecule encodes for several proteins.
- As it is produced from multiple genes, it contains numerous start and termination codons.
mRNA Synthesis – Transcription and Processing
- RNA is synthesised from DNA via the transcription process.
- A single DNA-dependent RNA polymerase catalyses the transcription of all kinds of RNA in prokaryotes.
- In this instance, mRNA does not require processing, therefore transcription and translation can occur simultaneously in the cytosol, where both processes occur.
- In eukaryotes, RNA polymerase II transcribes pre-mRNA. mRNA is synthesised in the nucleus as pre-mRNA or primary transcript. It is known as heterogeneous nuclear RNA or hnRNA.
- Following maturation, the mature mRNA is transferred to the cytosol and translated.
Transcription
- The process of synthesising RNA from DNA is known as transcription. The DNA-dependent RNA polymerase synthesises RNA in the 5′ to 3′ direction utilising DNA as a template.
- The DNA strand with polarity 3′ to 5′ serves as a template strand, whereas the DNA strand with polarity 5′ to 3′ is known as the coding strand.
- The transcribed RNA has the same nucleotide sequence as the coding strand.
- A DNA transcription unit consists of three major sections, and all references are relative to the coding strand.
- Promoter – located at the 5′ terminus of the structural gene.
- Structural gene – located between the promoter and the terminator.
- Terminator – located at the 3′-end of the structural gene.
Steps of Transcription
The transcription procedure involves three steps. The following are:
- Initiation – RNA polymerase interacts to the area of the promoter. It initiates transcription by unwinding the DNA double helix into a transcription bubble.
- Elongation – RNA polymerase continues to utilise complementary nucleoside triphosphate as substrates, and elongation of the DNA double helix proceeds. RNA polymerase transcribes RNA in the direction from 5′ to 3′.
- Termination – When transcription reaches the terminator region, it ceases and mRNA detaches from RNA polymerase.
Processing of mRNA
The freshly generated mRNA or initial transcript is referred to as hnRNA and must be processed to become mature mRNA. Post-transcriptional processing of the hnRNA yields mature mRNA. The transfer of functional mRNA from the nucleus. In the cytoplasm, translation or protein synthesis occurs.
The hnRNA includes both exons and introns, which are respectively coding and noncoding sections. It is spliced, capped, and terminated.
- Splicing – Introns are deleted from the parent transcript and exons are linked together during this procedure.
- Capping – In this process, the 5′-end of hnRNA is capped with an uncommon nucleotide, methyl guanosine triphosphate.
- Tailing – At the 3′-end, 200-300 adenylate residues are added during the tailing procedure. In addition to protecting RNA against degradation by exonucleases, the poly(A) tail is involved in the termination of transcription, transport of mRNA, and translation.
The nuclear pore complex transports the fully functioning mRNA to the cytoplasm, where it is translated into a polypeptide chain.
Translation of mRNA
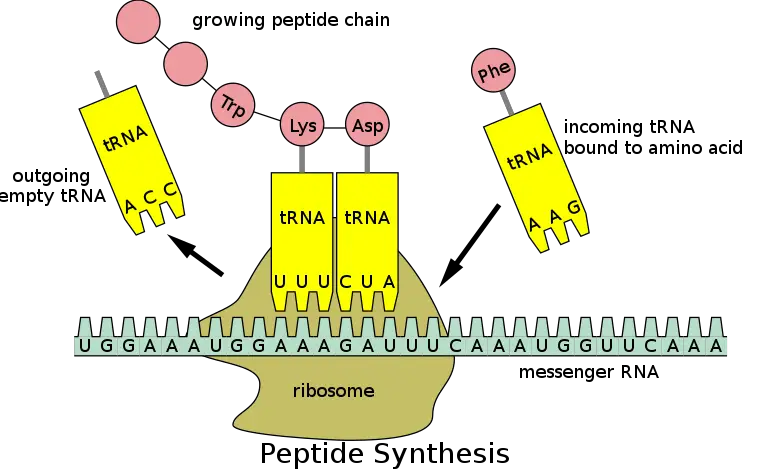
- With the aid of transfer RNA (tRNA) molecules and several proteins known as initiation, elongation, and termination factors, mRNA can be translated on unbound ribosomes in the cytoplasm.
- Proteins generated on unbound ribosomes in the cytoplasm are frequently utilised by the cell within the cytoplasm or within intracellular organelles.
- Alternately, proteins that must be secreted begin translation in the cytoplasm, but once the initial few residues are translated, specialised proteins transfer the whole translation machinery to the endoplasmic reticulum membrane (ER).
- The initial few amino acids are incorporated into the ER membrane, while the remainder of the protein is discharged into the ER’s inner region.
- The short sequence is eliminated from proteins that must be secreted from the cell, but proteins destined for intracellular membranes maintain the short stretch that serves as a membrane anchor.
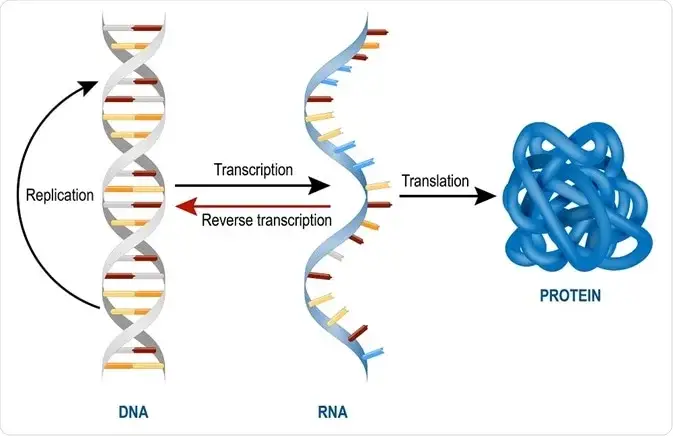
Circularization of mRNA
- In eukaryotes, mRNA molecules form circular shapes as a result of the interaction between eIF4E and poly(A)-binding protein, which bind to eIF4G to form an mRNA-protein-mRNA bridge.
- Circularization is believed to improve the cycling of ribosomes on mRNA, resulting in time-efficient translation, and may also serve to guarantee that only intact mRNA are translated (partially degraded mRNA characteristically have no m7G cap, or no poly-A tail).
- Other circularization methods occur, particularly in viral mRNA.
- The 5′ end of poliovirus mRNA contains a cloverleaf region that binds PCBP2, which in turn binds poly(A)-binding protein to form the typical mRNA-protein-mRNA circle.
- Without the involvement of proteins, the Barley yellow dwarf virus binds mRNA segments on its 5′ end and 3′ end (known as kissing stem loops), circularising the mRNA.
- RNA virus genomes, whose + strands are translated as messenger RNA, are also frequently circularised.
- During genome replication, circularization increases replication rates by cycling viral RNA-dependent RNA polymerase in a manner analogous to how the ribosome is thought to cycle.
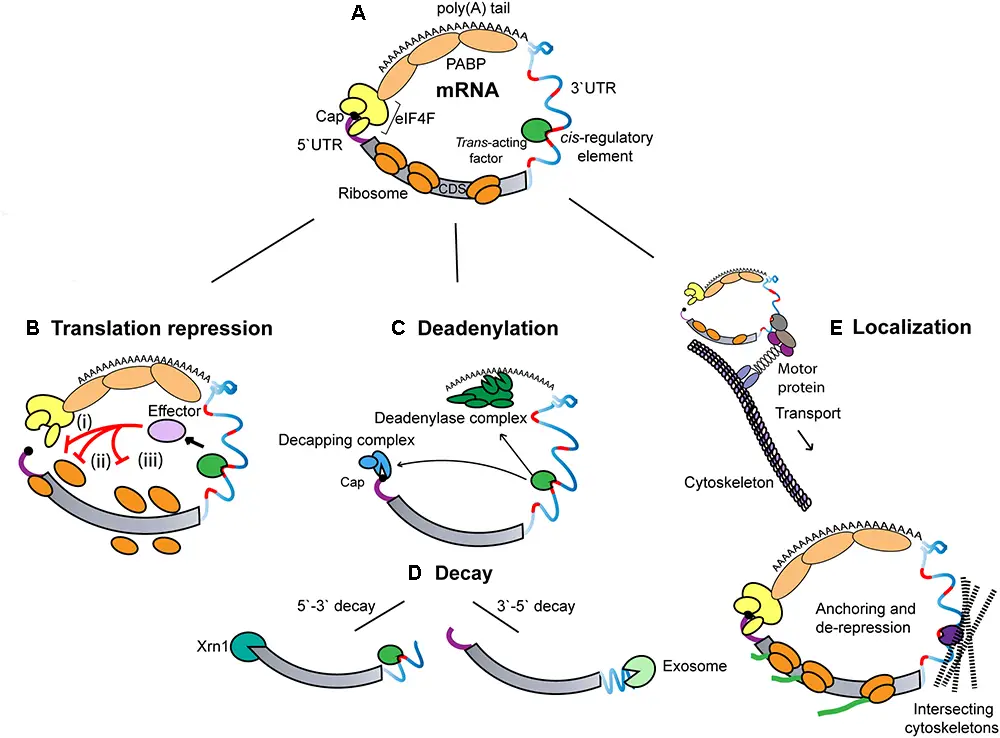
mRNA degradation
- Different mRNAs inside a same cell have different half-lives (stabilities).
- Individual mRNAs in bacterial cells can live for seconds to over an hour.
- The average lifetime of bacterial mRNA is between 1 and 3 minutes, making it far less stable than eukaryotic mRNA.
- mRNA lifetimes in mammalian cells varies from minutes to days.
- The more stable an mRNA is, the more protein that can be made from it.
- The short half-life of messenger RNA enables a cell to swiftly modify protein production in response to changing demands.
- Several of the mechanisms that result in the degradation of mRNA are outlined here.
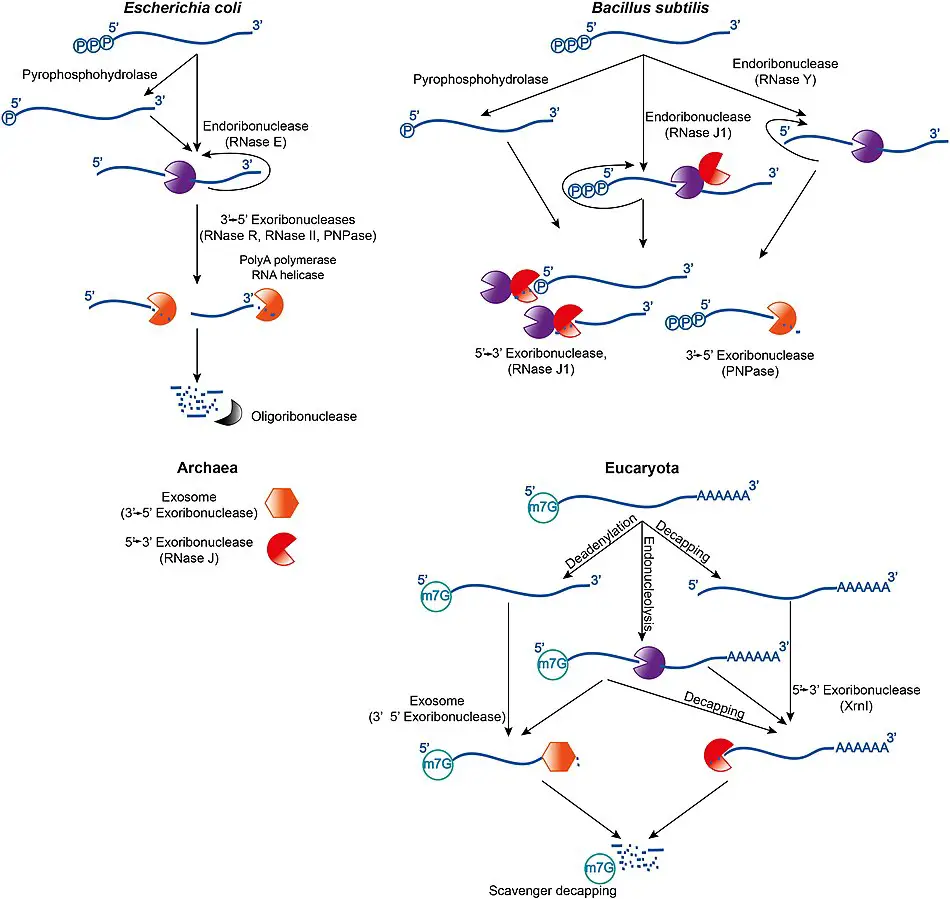
Prokaryotic mRNA degradation
- In general, the lifetime of mRNA in prokaryotes is significantly shorter than in eukaryotes.
- Prokaryotes utilise a mixture of ribonucleases, including endonucleases, 3′ exonucleases, and 5′ exonucleases, to degrade messages.
- Small RNA molecules (sRNA) that are tens to hundreds of nucleotides in length can sometimes increase the degradation of certain messenger RNAs (mRNAs) by base-pairing with complementary sequences and enabling ribonuclease cleavage by RNase III.
- Recent research indicates that bacteria have a 5′ cap comprised of a triphosphate at the 5′ end.
- Two of the phosphates are removed, leaving a 5′ monophosphate, which causes the message to be degraded by the exonuclease RNase J, which converts 5′ to 3′.
Eukaryotic mRNA turnover
- Inside eukaryotic cells, the processes of translation and mRNA decay work together in a way that keeps them in balance.
- ribosomes, the eukaryotic initiation factors eIF-4E and eIF-4G, and poly(A)-binding protein all bind to messages that are being actively translated. The ends of the message are protected because eIF-4E and eIF-4G block the decapping enzyme (DCP2) and poly(A)-binding protein blocks the exosome complex.
- The size and number of structures in the cytoplasm called P-bodies show how well translation and decay work together.
- Exonucleases that are directed to specific messenger RNAs by cis-regulatory sequences on the RNA and trans-acting RNA-binding proteins cut off the poly(A) tail of the messenger RNA.
- The removal of the Poly(A) tail is thought to break up the message’s circular structure and make the cap binding complex less stable.
- Then, either the exosome complex or the decapping complex breaks down the message.
- This makes it easy to get rid of messages that aren’t being used for translation, while active messages stay the same.
- We don’t fully understand how translation stops and the message is passed on to decay complexes.
AU-rich element decay
- When there are AU-rich elements in some mammalian mRNAs, cellular proteins that bind to these sequences and cause poly(A) tail removal tend to make these transcripts less stable.
- It is thought that when the poly(A) tail is lost, it makes it easier for both the exosome complex and the decapping complex to attack the mRNA.
- Rapid mRNA degradation through AU-rich elements is a key way to stop the overproduction of powerful cytokines like tumour necrosis factor (TNF) and granulocyte-macrophage colony-stimulating factor (GM-CSF).
- AU-rich elements also control how c-Jun and c-Fos, which are proto-oncogenic transcription factors, are made.
Nonsense-mediated decay
- nonsense-mediated decay (NMD), which looks for premature stop codons (nonsense codons) in eukaryotic messages, keeps an eye on eukaryotic messages.
- Incomplete splicing, V(D)J recombination in the adaptive immune system, DNA mutations, transcription mistakes, leaky scanning by the ribosome that causes a frame shift, and other things can cause these.
- When a premature stop codon is found, it causes mRNA to break down through 5′ decapping, 3′ poly(A) tail removal, or endonucleolytic cleavage.
Small interfering RNA (siRNA)
- Small interfering RNAs (siRNAs), which are made by Dicer, are a part of the RNA-induced silencing complex (RISC) in metazoans.
- This complex has an enzyme called an endonuclease that cuts apart messages that are perfectly complementary and that the siRNA binds to.
- Exonucleases then destroy the mRNA fragments that were made. siRNA is often used in labs to stop genes from working in cultures of cells.
- It is thought to be a part of the innate immune system that protects against viruses with two strands of RNA.
MicroRNA (miRNA)
- MicroRNAs (miRNAs) are small RNAs that are similar to sequences in messenger RNAs from metazoans.
- When a miRNA binds to a message, it can stop the message from being translated and speed up the removal of the poly(A) tail, which speeds up mRNA degradation.
- Researchers are still trying to figure out how miRNAs do what they do.
Other decay mechanisms
- Messages can also be broken down in other ways, such as by nonstop decay or by Piwi-interacting RNA (piRNA) that makes them silent.
Processing of mRNA in Prokaryotes and Eukaryotes
- mRNA transcripts undergo minimal or no processing in prokaryotes. 5′-terminal degradation of prokaryotic mRNA is highly fast.
- Therefore, to prevent deterioration, it is translated before to transcription. Ribosomes assemble on incomplete mRNA, and the first cistron (protein-coding region) is rapidly translated.
- The internal cistrons are partially protected by the 5′ and 3′ stem-loop structures.
- Different genes are transcribed from snRNA genes by eukaryotic RNA Pol II, resulting in heterogeneous nuclear RNA (hnRNA). Transcripts of pre-mRNA undergo processing to generate mature mRNAs.
- The following processing events are briefly described:
1. The hnRNP
- RNA Pol II produces hnRNA, which is predominantly composed of pre-mRNA. A, B, and C proteins) to form heterogeneous nuclear ribonucleoprotein (hnRNP) particles shortly after production of hnRNA.
- They include three copies of three tetramers and between 600 and 700 hnRNA nucleotides.
- It is possible that hnRNP proteins maintain the hnRNA in a single-stranded state and aid in certain RNA processing activities.
2. The snRNP Particles
- Numerous uracil-rich snRNA molecules, marked U1, U2, etc., exist. The majority of them are transcribed by RNA Pol II, which forms snRNPs with particular proteins.
- U1, U2, U4, U5, and U6 are the most common proteins involved in pre-mRNA splicing.
- The majority of them determine the pre-rRNA methylation sites and are positioned in the nucleolus.
- The main snRNAs with the sequence 5′-RA(U)n GR-3′ bind with eight cytoplasmic proteins and become hypermethylated. After that, they are returned to the nucleus.
3. 5′-Capping
- Immediately after RNA pol II synthesises an approximately 25-nucleotide-long mRNA chain, the 5′-end is chemically changed by the incorporation of a 7-methylguanosine (m7G) residue.
- This 5′ change is referred to as a CAP and is performed by adding a GMP nucleotide to the new transcript.
- It is inserted with a reverse polarity (5′ to 5′ triphosphate bridge) compared to the typical linkage (by an enzyme mRNA guanyltransferase).
- This cap inhibits 5′-exonuclease attack, but facilitates splicing transport and translation.
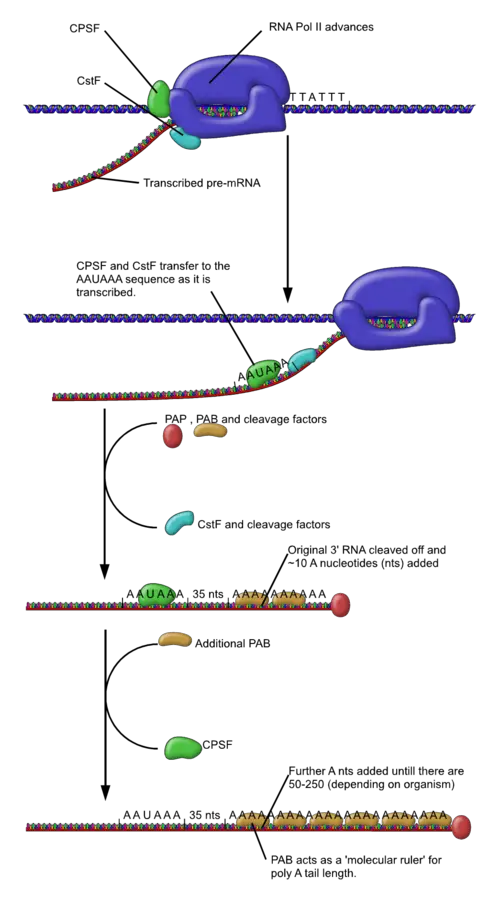
4. 3′ Cleavage and Polyadenylation
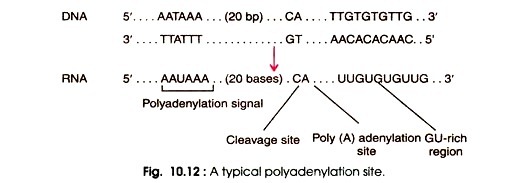
- The 3′-end of the majority of pre-mRNA molecules is formed through cleavage during polyadenylation.
- For cleavage and polyadenylation reactions, DNA and its pre-mRNA transcript containing the polyadenylation signal 5′-AAUAAA-3′ contain particular sequences.
- It is followed by a GU-rich 5′-YA-3′ sequence. These sequences are known together as polyadenylation sites.
- Then, poly (A) polymerase (PAP) adds an approximately 250-nucleotide poly (A) tail and mature mRNA is produced.
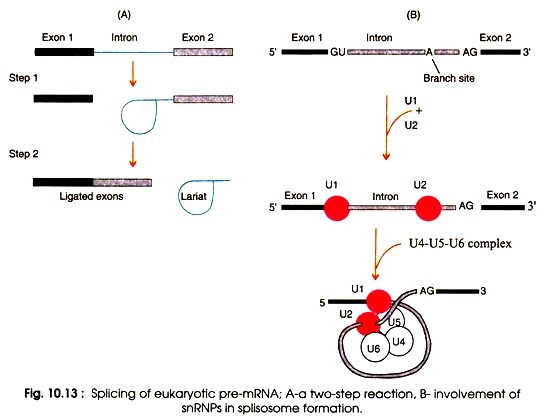
5. Splicing
- The exons of eukaryotic pre-mRNA are interrupted by introns (intervening sequencing) (the coding regions).
- The pre-mRNA is spliced, the introns are eliminated, and two flanking exons are linked. This procedure is known as splicing.
- This occurs in the nucleus prior to the transfer of mature mRNA into the cytoplasm. Introns require a 5′-GU, an AG-3′, and a branch-point sequence for splicing.
- In a two-step procedure, the introns are extracted as lariat, a circular molecule with a tail that is destroyed later.
- The process of splicing is facilitated by the snRNPs (U1, U2, U4, U5, and U6) and other splicing factors.
- The complex of snRNPs and pre-mRNA that holds the upstream and downstream exons close together is known as the splisosome. The looping out introns are also known as splisomes.
- Inside the splisosome, cleavage of introns and ligation of exons occur, releasing the introns as a lariat.
6. Pre-mRNA Methylation
- A small proportion of A residues in pre-mRNA (bearing the sequence 5′-RRACX-3′, where R = purine) are methylated at the N6 position.
Disorders Related to mRNA Processing
- Over 200 diseases are linked to problems with how pre-mRNA is turned into mRNA.
- Changes in DNA or the machinery that does the splicing have a big effect on how well pre-mRNA is spliced. For instance, a wrong DNA sequence can get rid of, weaken, or turn on hidden splice sites in pre-mRNA.
- Also, if the machinery for splicing isn’t working right, the spliceosome may cut the pre-mRNA in the wrong place, no matter what the sequence is. Because of these mutations, pre-mMRA is turned into mRNAs, which are then used to make proteins that don’t work right.
- Sometimes, nonsense-mediated mRNA decay and co-transcriptional degradation of new pre-mRNAs go after the abnormal mRNAs themselves.
- Cells from people with diseases like progeria, breast cancer, and cystic fibrosis have RNA splicing problems. Cancer and neuropathological diseases are the most common types of these problems.
Difference between Prokaryotic mRNA vs. Eukaryotic mRNA
- Polycistronic prokaryotic mRNAs have more than one place where protein synthesis can start and stop. Eukaryotes only have one place where translation starts, and most of their mRNAs are monocistronic.
- Since prokaryotes don’t have organelles or a clear nuclear envelope, mRNA transcription and translation can happen at the same time in the cytoplasm. In eukaryotes, chromosomes in the nucleus transcribe mRNA, which is then processed and moved through nuclear pores into the cytoplasm.
- In eukaryotes, translation doesn’t happen until transcription is done, which is different from prokaryotes.
- Ribonucleases, which cut RNA, are always cutting up prokaryotic mRNA. For example, mRNA in E. Coli has a half-life of about two minutes. Bacterial mRNAs don’t last long so that bacteria can respond quickly to changes in their environment.
- Eukaryotic mRNAs are more stable when it comes to metabolism. For example, reticulocytes, which are the precursors of red blood cells in mammals, make haemoglobin by translating mRNAs that were made when the nucleus was still there. This process takes several days.
- Lastly, prokaryotes don’t do much processing to their mRNAs. In eukaryotes, the pre-mRNA must be processed before it can be translated. This means that the introns must be taken out and the 5′-cap and 3′ poly-adenylated tail must be added. Only then is the mature mRNA ready to be translated.
Functions of Messenger RNA (mRNA)
- Template for Protein Synthesis: The main job of mRNA is to serve as a blueprint for making proteins. Ribosomes that are free in the cytoplasm or on the rough endoplasmic reticulum can do the translation (RER).
- Genetic information: The genetic information in the RNA is turned into amino acids in the polypeptide chain. The order of amino acids in a polypeptide chain is set by the order of nucleotides or codons in the mRNA. Each triplet codon, which is a group of three nucleotides, tells the cell to make a certain amino acid. There can be more than one codon for an amino acid. Codons do not overlap.
- Protein Synthesis: mRNA tells the cell how to make a protein. mRNA sequences that have been changed can be used to treat diseases and also as a vaccine. Several mRNA vaccines have been made and are being worked on to protect against COVID-19 infection. Some of these vaccines, like the Pfizer-BioNTech and Moderna vaccines, have been approved for limited use.
- Complementary DNA library: mRNAs have made it possible to make a library of complementary DNA.
- Therapeutic drugs: mRNA-based therapeutics are starting to look like a good type of medicine. Some COVID vaccines now use this mRNA technology instead of the attenuated virus to make the patient’s immune system react.
References
- Shyu AB, Wilkinson MF, van Hoof A. Messenger RNA regulation: to translate or to degrade. EMBO J. 2008;27(3):471-481. doi:10.1038/sj.emboj.7601977.
- Bentley, D. Coupling mRNA processing with transcription in time and space. Nat Rev Genet 15, 163–175 (2014). https://doi.org/10.1038/nrg3662.
- Wang D, Farhana A. Biochemistry, RNA Structure. [Updated 2021 May 9]. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2021 Jan-. Available from: https://www.ncbi.nlm.nih.gov/books/NBK558999/.
- https://openstax.org/books/microbiology/pages/10-3-structure-and-function-of-rna
- https://www.britannica.com/science/messenger-RNA
- https://biologydictionary.net/mrna/
- https://www.news-medical.net/life-sciences/-Types-of-RNA-mRNA-rRNA-and-tRNA.aspx
- https://www.genome.gov/genetics-glossary/RNA-Ribonucleic-Acid
- https://en.wikipedia.org/wiki/Messenger_RNA
- https://www.cd-genomics.com/blog/mrna-fact-sheet-definition-structure-function-and-association-with-disease/
- https://www.genome.gov/genetics-glossary/messenger-rna
- https://thebiologynotes.com/messenger-rna/
