Table of Contents
A phylogenetic tree (also phylogeny or evolutionary tree [3]) is a branching diagram or a tree that illustrates the evolutionary relationships between various biological species or other entities based on similarities and differences in their physical or genetic characteristics. All life on Earth belongs to a singular phylogenetic tree, indicating a shared ancestry.
In a rooted phylogenetic tree, each node with descendants represents the inferred most recent common ancestor of those descendants[4], and in some trees, the edge lengths can be interpreted as time estimates. Every node is referred to as a taxonomic unit. Internal nodes are typically referred to as hypothetical taxonomic units because they cannot be observed directly. Biomedical disciplines such as bioinformatics, systematics, and phylogenetics find trees valuable. Unrooted trees illustrate only the relatedness of the leaf nodes and do not necessitate the knowledge or inference of the ancestral root.
Ancient conceptions of a ladder-like progression from lower to higher forms of life (such as in the Great Chain of Being) gave rise to the concept of a tree of life. Early depictions of “branching” phylogenetic trees include a “paleontological chart” depicting the geological relationships between plants and animals in Edward Hitchcock’s (1800s) book Elementary Geology.
In his 1859 book On the Origin of Species, Charles Darwin depicted an evolutionary “tree” as a diagram. Over a century later, evolutionary biologists continue to use tree diagrams to illustrate evolution because they effectively convey the concept that speciation occurs through the adaptive and semirandom division of lineages.
The term phylogenetic or phylogeny is derived from the ancient Greek words v (phûlon), which means “race” or “lineage,” and (génesis), which means “origin” or “source.”
What is a phylogenetic tree? – Phylogenetic tree definition
- A phylogenetic tree, also referred to as an evolutionary tree or a tree of life, is a branching diagram that depicts the evolutionary relationships between various species or groups of organisms. It illustrates the evolutionary history and genetic relationships between organisms, highlighting their common ancestry and the branching patterns of their evolutionary divergence.
- The structure of a phylogenetic tree reflects the relationship between organisms based on shared traits, genetic data, and other evidence. It is composed of nodes, which represent hypothetical common ancestors, and branches, which represent evolutionary trajectories and species relationships. The arrangement and length of the branches indicate the amount of evolutionary change that has occurred over time, with longer branches indicating greater divergence.
- A variety of data sources, including comparative anatomy, molecular genetics, and fossil documents, are used to construct phylogenetic trees. Researchers analyze shared characteristics and genetic sequences to infer evolutionary relationships and determine which organisms are more closely related and which diverged earlier in the course of evolution. A phylogenetic tree is constructed using statistical methods and algorithms that analyze the data and generate the most plausible branching patterns.
- Our current comprehension of the relationships between organisms is represented by these trees, which are neither fixed nor definitive. As new data and discoveries become available, the structure of phylogenetic trees may be revised and updated, resulting in a more refined understanding of the evolutionary relationships between species.
- Phylogenetic trees provide valuable insights into the patterns and processes of evolution, allowing scientists to investigate the origins of species, the diversification of life forms, and the mechanisms underlying evolutionary change. They are fundamental to many disciplines, including evolutionary biology, ecology, taxonomy, and conservation, and provide a framework for comprehending the interconnectedness and diversity of life on Earth.
Parts of a Phylogenetic Tree
A phylogenetic tree depicts the evolutionary connections between various organisms or species. It is a diagram depicting the common ancestor and divergence of species over time. The primary components of a phylogenetic tree are as follows:
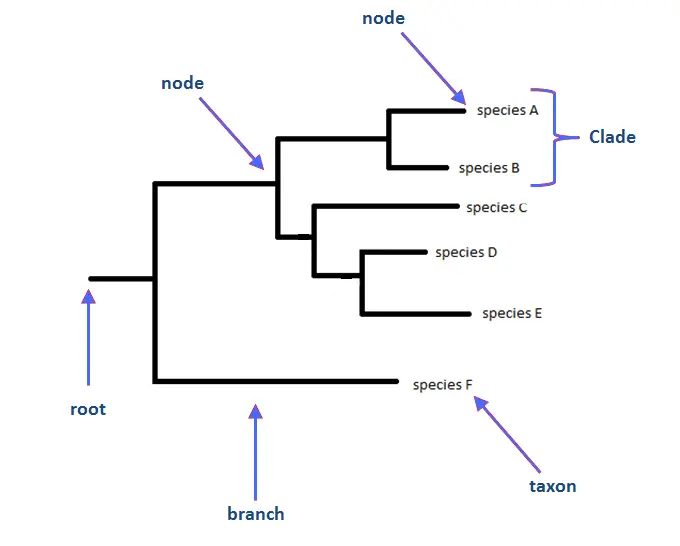
- Branches: The branches of a phylogenetic tree represent evolutionary lineages or the descent line. They connect the nodes or divergence sites.
- Nodes: Nodes represent a common ancestor of the species or groups that diverged from it. Nodes can be referred to as internal nodes or internal branches when they represent non-observable common ancestors.
- Tips or Leaves: The tips or leaves of a phylogenetic tree represent extant or alive species or groups. Terminal taxa are taxa that are located at the extremities of branches.
- Root: The root of the phylogenetic tree represents the most recent common ancestor of all the included species or groups. Typically, it is depicted at the base of the tree.
- Branch Length: In a phylogenetic tree, the branch length represents the quantity of evolutionary change that has occurred along a particular branch. In terms of time (e.g., millions of years) or genetic variation (e.g., DNA substitutions), the duration can be quantified.
- Phylogenetic Distance: The phylogenetic distance between two species or groups quantifies how closely they are related. Typically, it is estimated using genetic or morphological differences.
- Taxa: Taxa are the categories of organisms or species that comprise the phylogenetic tree. Individual species to higher taxonomic levels such as genera, families, orders, and even larger groups.
- Clades: Clans are monophyletic entities in a phylogenetic tree, composed of an ancestor and all of its descendants. They share unique characteristics that are derived from a common ancestor.
It is essential to observe that phylogenetic trees can be represented in a variety of ways, including cladograms, phylograms, and chronograms, each of which emphasizes a different aspect of evolutionary relationships or evolutionary time.
Types of Phylogenetic Tree
On the basis of the presence or absence of a common root
On the premise of the presence or absence of a common ancestor, phylogenetic trees can be divided into two distinct categories:
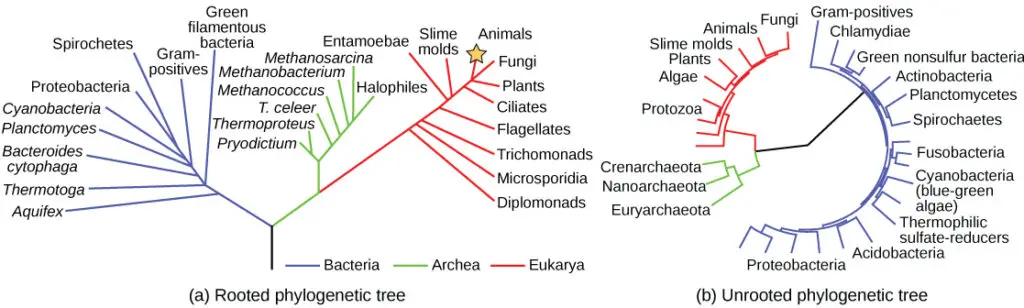
1. Rooted Phylogenetic Tree
- Rooted phylogenetic trees illustrate the evolutionary relationships between taxa that share a common ancestor.
- The root represents the most recent ancestor shared by all taxa in the tree. It is typically positioned at the base of the tree and functions as a point of reference for interpreting branching patterns and evolutionary relationships among taxa.
- The directionality of evolution and the relative timeline of divergence events are revealed by rooted trees.
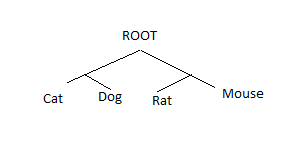
2. Unrooted Phylogenetic Tree
- Unrooted phylogenetic trees depict the evolutionary relationships between taxa that lack a designated common root.
- Instead of a singular root, unrooted trees depict taxonomic relationships as a series of connected branches and nodes. Unrooted trees cannot provide information about the direction of evolution or the relative timeline of divergence events because they lack a root.
- Unrooted trees are primarily used to visualize the relationships between taxa, but they can also be utilized to infer evolutionary patterns and identify clusters or groups of closely related taxa.
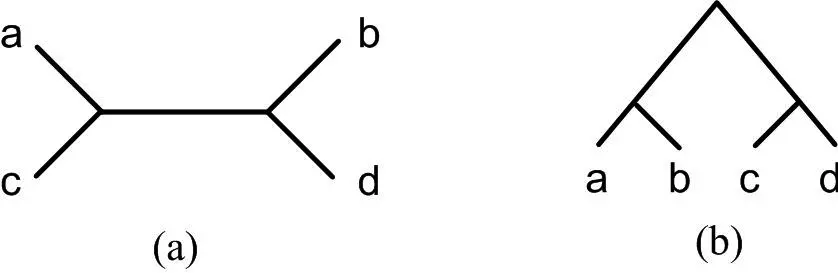
Both rooted and unrooted phylogenetic trees offer unique benefits and applications. When the relative timing of divergence events and the direction of evolution are essential, rooted trees are frequently employed. They are particularly valuable for studying the history of evolution and deducing ancestral states. In contrast, unrooted trees are frequently used for exploratory analysis, clustering analysis, and initial visualization of taxonomic relationships. When the concentration is on the relative relationships between taxa as opposed to their absolute evolutionary history, they are particularly useful.
Types of Phylogenetic Tree On the basis of topology
On the basis of their topology, phylogenetic trees can be divided into the following categories:
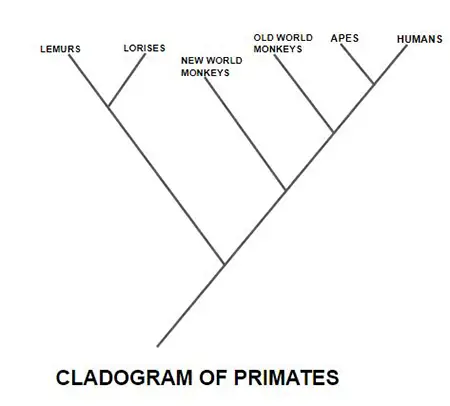
1. Cladogram
A cladogram is a form of phylogenetic tree that illustrates the branching pattern of evolutionary relationships between taxa. It indicates the order of divergence but not the quantity of time or evolutionary change that has taken place. Cladograms consist of branches that divide at nodes, representing common ancestors, and their lengths are typically equal.
2. Phylogram
A phylogram is a form of phylogenetic tree that uses branch lengths to represent the degree of evolutionary change. Indicating the relative amount of time or evolutionary distance between nodes, branch lengths can be proportional to genetic or morphological differences. Phylograms add to our understanding of the magnitude of evolutionary change.
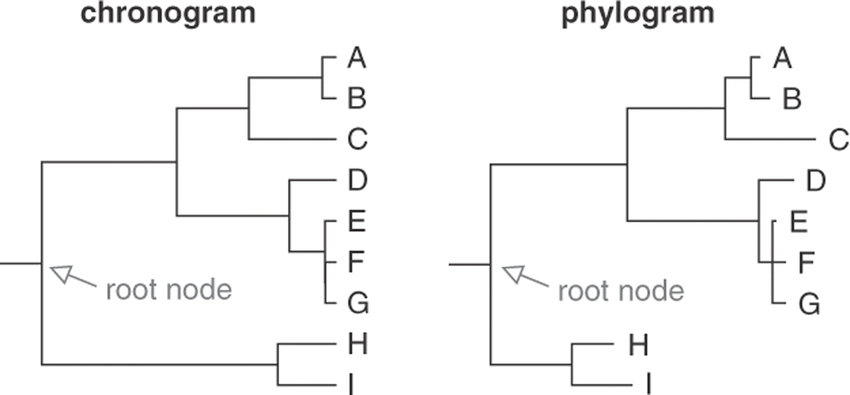
3. Chronogram
A chronogram, also known as a time tree, is a scaled phylogenetic tree that depicts the estimated time of divergence between species or groups. In a chronogram, the branch lengths are proportional to the estimated time since divergence, which is typically derived from fossil records or molecular clock analyses. Chronograms provide a temporal perspective, enabling the visualization of evolutionary event timing.
4. Network Tree
A network tree, also known as a reticulated tree, is a form of phylogenetic representation consisting of reticulations or interconnected branches. When complex evolutionary processes, such as hybridization, horizontal gene transfer, or recombination, cannot be adequately represented by a simple bifurcating tree structure, this method is employed. The exchange of genetic material between distinct lineages can be visualized via network structures.
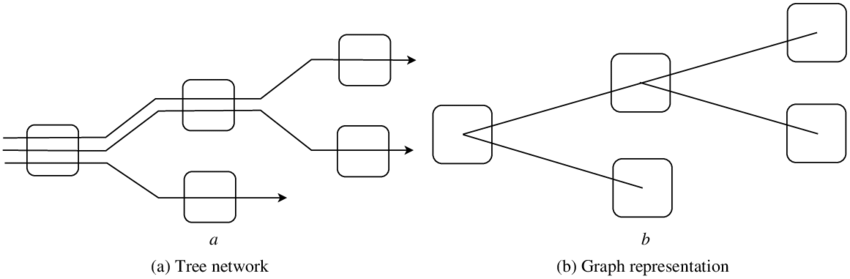
5. Fan Tree
A fan tree is a form of phylogenetic tree that illustrates the relationships between taxa by utilizing multiple branches emanating from a single node. It is frequently used to represent rapid radiations or evolutionary events in which multiple lineages emerge from a common ancestor within a brief period of time. Fan trees illustrate the divergence of lineages without specifying their sequence.
6. Polytomy
A polytomy is a characteristic of a phylogenetic tree in which three or more branches arise from a single node. It signifies an unresolved relationship in which the precise order or sequence of divergence is unknown. Polytomies can occur when there is insufficient data or when simultaneous rapid divergence events occur.
Differences Between Chronogram and Phylogram tree
tree:
| Criteria | Chronogram Tree | Phylogram Tree |
|---|---|---|
| Time Information | Includes time scale, indicating the estimated time of divergence between lineages. | Does not include explicit time scale information. |
| Branch Lengths | Branch lengths represent evolutionary time (usually in units such as years or generations). Longer branches indicate longer periods of evolutionary divergence. | Branch lengths typically represent some measure of genetic or evolutionary distance. They do not directly represent time. |
| Time Calibration | Requires calibration points (fossil records, geological events) to estimate the time scale and assign absolute ages to internal nodes. | Does not require explicit time calibration. |
| Inference Method | Often constructed using molecular clock methods that estimate the rate of molecular evolution and use it to infer divergence times. | Can be constructed using various methods such as maximum likelihood, maximum parsimony, or distance-based methods. |
| Applications | Useful for studying evolutionary patterns, rates of molecular evolution, and estimating divergence times. | Commonly used for studying phylogenetic relationships, inferring ancestral states, and identifying evolutionary patterns. |
| Visual Representation | Often displayed with horizontal or diagonal lines indicating the time axis along the branches. | Typically displayed with branches of equal length, creating a symmetrical tree representation. |
| Example | A chronogram might show the estimated divergence times of different primate species, indicating when they branched off from a common ancestor. | A phylogram might show the genetic distances between different bird species, illustrating their evolutionary relationships based on genetic similarities. |
Phylogenetic Tree Construction Steps/How to Make a Phylogenetic Tree?
Phylogenetic tree construction requires multiple stages and considerations. Here is an overview of the procedure:
- Define the Investigational Question: Determine your phylogenetic analysis’s purpose. Are you curious about the evolutionary relationships between species, populations, and genes? Clarify the scope and intent of your research.
- Data Collection: Collect the required genetic or morphological information for the taxa or sequences of interest. This may involve sequencing DNA or gathering sequence information from public databases. Ensure that the data is accurate, representative, and properly aligned.
- Sequence Alignment: Align the sequences using the appropriate software or instruments for alignment. Sequence alignment assures that homologous positions within the sequences are accurately matched, allowing for precise comparison and analysis.
- Select a Phylogenetic Method: Based on your data type, research query, and available computational resources, select an appropriate method for tree construction. Distance-based methods (e.g., Neighbor-Joining), character-based methods (e.g., Maximum Likelihood, Maximum Parsimony), and Bayesian Inference are common statistical techniques. Each method has its own assumptions, advantages, and disadvantages.
- Choose a Substitution Model: If using character-based methods, choose a substitution model that accurately describes the evolution of your data. Various models account for various evolutionary factors, including nucleotide/amino acid substitutions, rate heterogeneity, and variation between sites. The model should reflect the most plausible scenario of evolution for your data.
- Tree Construction: Utilize the selected technique or software to construct the phylogenetic tree. The software will use algorithms and statistical calculations to estimate the tree topology and branch lengths that provide the best explanation for the evolutionary relationships between the sequences or taxa.
- Evaluate Tree Robustness: Evaluate the tree’s robustness by conducting statistical tests and generating support values (e.g., bootstrap values, posterior probabilities) via resampling or Bayesian analyses. larger confidence in the inferred relationships is indicated by larger support values.
- Tree Visualization: Once the tree has been constructed, it should be visualized using specialized software or tree visualization tools. For clarity and interpretation, graphically represent the tree with the appropriate labels, color coding, and annotations.
- Interpretation and Analysis: In the context of your research query, interpret the phylogenetic tree. Analyze the relationships between the taxa or sequences, and identify monophyletic groups, evolutionary trends, and divergence patterns. If applicable to your research, consider additional analyses such as ancestral state reconstruction and divergence time estimation.
- Refinement through Iteration: Phylogenetic analysis is an iterative process. Critically evaluate the results, consider alternative tree-building methodologies or models, and if necessary, refine the analysis. To ensure accurate and reliable conclusions, reevaluate data quality, alignment, and any possible biases or limitations.
It is essential to note that the procedure may vary based on the particular research objectives, data type, and available resources. You can also be guided through the specific details and considerations for your phylogenetic analysis by consulting relevant literature, textbooks, or seeking expert advice.
Phylogenetic Tree Construction Methods
The two main methods for constructing phylogenetic trees are the distance-based method and the character-based method.
1. Distance-Based Methods
Distance-based methods construct phylogenetic trees based on the genetic or morphological distances between taxa. These methods use pairwise comparisons of sequences or traits to calculate the distances and then build the tree based on these distances. Common distance-based methods include Neighbor-Joining (NJ) and Unweighted Pair Group Method with Arithmetic Mean (UPGMA).
Steps involved in distance-based tree construction
- Calculate pairwise distances: Compute the genetic or morphological distances between all pairs of taxa based on sequence alignment or trait comparisons.
- Build a distance matrix: Construct a matrix that represents the pairwise distances between taxa.
- Tree construction: Use algorithms such as Neighbor-Joining or UPGMA to build a tree based on the distance matrix. These algorithms iteratively join the closest pairs of taxa to form the tree, with branch lengths proportional to the distances.
Different Methods of Distance-Based
Two commonly used distance-based methods for constructing phylogenetic trees are Neighbor-Joining (NJ) and Unweighted Pair Group Method with Arithmetic Mean (UPGMA).
- Neighbor-Joining (NJ): Neighbor-Joining is a distance-based method that constructs phylogenetic trees by iteratively joining the closest pairs of taxa based on their genetic or morphological distances. It aims to minimize the total branch length of the tree. The NJ algorithm starts with a star-like tree and repeatedly joins the pair of taxa with the smallest pairwise distance until a complete tree is formed.
- Unweighted Pair Group Method with Arithmetic Mean (UPGMA): UPGMA is another distance-based method for constructing phylogenetic trees. It assumes a constant rate of evolution or a molecular clock. UPGMA builds the tree by iteratively merging the two closest taxa or groups of taxa based on their pairwise distances, using an average distance calculation. It produces a binary tree with branch lengths representing the average genetic or morphological distances.
Both Neighbor-Joining and UPGMA are widely used distance-based methods due to their simplicity and computational efficiency. These methods are particularly suitable when there is a need for quick estimation of trees, such as in large-scale datasets or when a molecular clock assumption is appropriate. However, it’s important to note that distance-based methods may be sensitive to violations of the molecular clock assumption and can produce biased results in certain scenarios.
Researchers often compare the results obtained from distance-based methods with other methods, such as character-based methods like Maximum Likelihood and Maximum Parsimony, to assess the robustness and accuracy of the inferred phylogenetic relationships.
2. Character-Based Methods
Character-based methods construct phylogenetic trees based on the analysis of specific characters or traits. These methods use the presence or absence of shared derived traits (synapomorphies) to infer evolutionary relationships. Character-based methods include Maximum Parsimony (MP), Maximum Likelihood (ML), and Bayesian Inference (BI).
Steps involved in character-based tree construction
- Character data collection: Collect and compile the presence or absence of specific characters or molecular sequence data for each taxon.
- Character matrix creation: Construct a matrix that represents the presence or absence of characters for each taxon.
- Tree construction: Use methods like Maximum Parsimony, Maximum Likelihood, or Bayesian Inference to find the tree topology that best explains the observed character data. These methods aim to minimize the number of character-state changes or maximize the likelihood of the observed data under a specific model of evolution.
Different Methods of Character-Based
Character-based methods include Maximum Parsimony (MP), Maximum Likelihood (ML), and Bayesian Inference (BI). These methods are widely used in phylogenetic tree construction and inference based on character data.
- Maximum Parsimony (MP): Maximum Parsimony aims to construct a phylogenetic tree by minimizing the number of evolutionary changes required to explain the observed character data. It assumes that the tree with the fewest character-state changes is the most likely or represents the true evolutionary history. MP methods search for the tree topology that minimizes the total number of character-state changes.
- Maximum Likelihood (ML): Maximum Likelihood estimates the most likely tree topology and model parameters given the observed character data and a specific model of evolution. ML methods calculate the likelihood of the observed data under different tree topologies and model parameters and search for the tree that maximizes this likelihood. ML methods can incorporate complex models of sequence evolution and can account for rate heterogeneity among sites.
- Bayesian Inference (BI): Bayesian Inference constructs phylogenetic trees by sampling from the posterior probability distribution of tree topologies and model parameters. It uses Bayesian statistical methods to estimate the relative probabilities of different tree topologies. BI incorporates prior information and likelihood calculations to generate a posterior distribution of trees. This allows for the quantification of uncertainty in tree inference.
These character-based methods differ in their underlying assumptions, computational approaches, and statistical frameworks. Each method has its strengths and limitations, and the choice of method depends on factors such as the available data, the complexity of the evolutionary process, and the desired level of statistical inference. Researchers often compare the results from multiple methods to assess the robustness of the inferred phylogenetic relationships.
Both distance-based and character-based methods have their advantages and limitations. The choice of method depends on factors such as the available data, computational resources, and the specific research question at hand. It is common to perform multiple analyses using different methods to assess the robustness of the inferred phylogenetic relationships.
| Method | Approach | Advantages | Limitations |
|---|---|---|---|
| Distance-Based Methods | Construct trees based on genetic distances or morphological similarities between taxa | Computational efficiency, suitable for large datasets, can handle diverse data types | Sensitive to violations of the molecular clock assumption, may produce biased results in certain scenarios |
| – Neighbor-Joining (NJ) | Iteratively joins closest pairs of taxa based on pairwise distances | Quick estimation, suitable for large datasets | May not always recover the true tree topology |
| – Unweighted Pair Group Method with Arithmetic Mean (UPGMA) | Merges closest taxa/groups based on average pairwise distances | Quick estimation, assumes a constant rate of evolution | Assumes a molecular clock, sensitive to unequal rates of evolution |
| Character-Based Methods | Infer evolutionary relationships based on shared derived traits (synapomorphies) | Can incorporate complex models of evolution, can handle violations of the molecular clock assumption | Computationally intensive, may be sensitive to model assumptions, may require large amounts of data |
| – Maximum Parsimony (MP) | Minimizes the number of evolutionary changes required to explain the observed data | Simplicity, intuitive interpretation | May be sensitive to long branch attraction, lacks statistical framework |
| – Maximum Likelihood (ML) | Estimates the most likely tree topology and model parameters given the observed data | Statistical rigor, can incorporate complex evolutionary models | Computationally intensive, model selection can be challenging |
| – Bayesian Inference (BI) | Samples from the posterior probability distribution of trees given the data and prior information | Accounts for uncertainty, incorporates prior information | Computationally intensive, requires specifying prior distributions |
How to Read a Phylogenetic Tree?
Reading Phylogenetic trees is identical to reading family trees. However, we will explain in detail how to read a Phylogenetic tree.
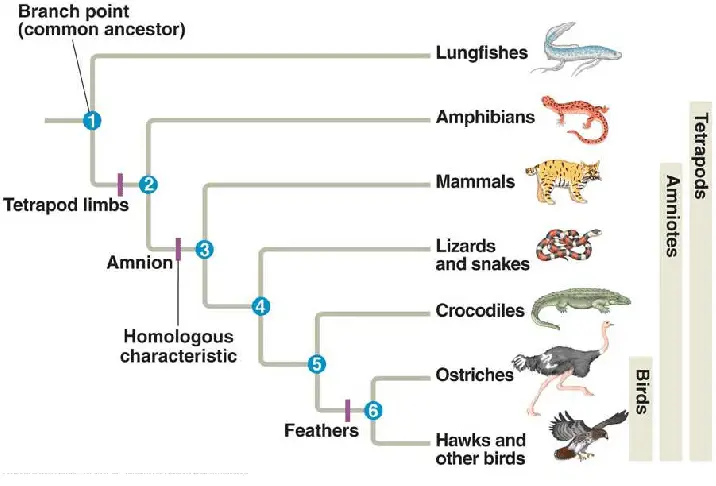
Step 01: See the Root as Ancestor and Tips as Descendants
The root of the tree represents the ancestor from which all species have evolved. And the apex represents the species group (a portion of their common ancestry).
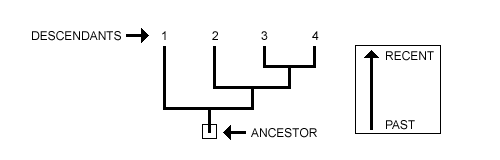
Step 02: See the Daughter Lineages
The second step is to identify the offspring lineages descended from the common ancestor. It divides into two or more branches that serve as a dividing line between one or more species.
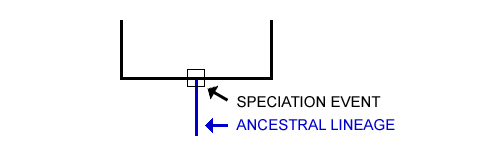
Step 03: See the Shared Ancestry Between Lineages (If Any)
The third stage is to determine whether a group of species share any ancestry lineages. See points B and C in the following diagram. Point B and C have unique histories, unlike Point A.
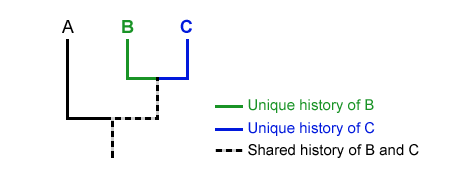
Step 04: Read the Diagram as a Whole
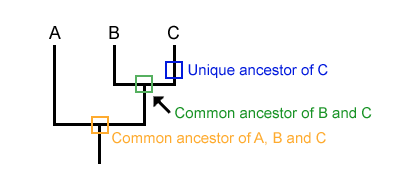
See the figure below, and you should now be able to comprehend it in its entirety. The orange color indicates that Points A, B, and C share a common ancestor. However, the Green color indicates that Points B and C share a unique but common ancestor, and the same is true of Point C.
Importance of Phylogenetic Tree/Applications of the phylogenetic tree
Phylogenetic trees are crucial to numerous areas of biological research and play several crucial roles:
- Understanding Evolutionary Relationships: Phylogenetic trees depict the evolutionary relationships between organisms. They aid in the comprehension of patterns of common ancestry, divergence, and evolutionary history. By examining phylogenetic trees, we can determine the evolutionary ancestry of various species or groups and determine their relatedness.
- Classification and Taxonomy: Phylogenetic trees serve as the foundation for classification and taxonomy. They provide a framework for classifying organisms into hierarchical categories according to their evolutionary relationships. By analyzing the branching patterns and common ancestry depicted in phylogenetic trees, scientists are able to assign species to the appropriate taxonomic ranks and determine their systematic placement.
- Testing Hypotheses: Phylogenetic trees enable scientists to formulate and test hypotheses about evolutionary processes such as speciation, extinction, adaptation, and biogeography. By comparing various tree topologies and assessing statistical support, scientists are able to evaluate competing hypotheses and draw conclusions about the evolutionary mechanisms that have shaped biodiversity.
- Biomedical and Medical Research: In biomedical and medical research, phylogenetic trees are useful for understanding the evolutionary origins and relationships of pathogens, parasites, and diseases. Researchers can monitor the spread of infectious diseases, identify the source of outbreaks, and study the evolution of drug resistance by constructing phylogenetic trees from genetic data. Phylogenetic trees also aid in the identification of potential zoonotic diseases and the forecasting of their future transmission patterns.
- Conservation and Biodiversity: Phylogenetic trees contribute to conservation efforts by revealing evolutionary uniqueness and species relationships. They aid in the identification of species that are evolutionarily distinct or at risk of extinction. Metrics of phylogenetic diversity can guide conservation prioritization by highlighting lineages that represent a substantial quantity of evolutionary history and may require special conservation attention.
- Comparative Genomics and Functional Studies: In comparative genomics, phylogenetic trees are used to infer ancestral states and reconstruct the evolution of genes and genomes. By analyzing branching patterns and patterns of gene presence/absence, researchers can identify genes that have been gained or lost during evolution and investigate their functional implications.
- Understanding Trait Evolution: Phylogenetic trees permit researchers to examine the evolution of traits across species. By mapping particular traits or characteristics onto a phylogenetic tree, scientists are able to analyze patterns of trait evolution, identify convergent or divergent evolution, and investigate the factors influencing phenotypic diversity.
In general, phylogenetic trees are a useful instrument for comprehending the evolution, relationships, and diversity of life on Earth. They provide a framework for the study of evolutionary processes, the organization of biodiversity, and the investigation of a vast array of biological questions across disciplines.
Phylogenetic tree maker Tool
There are several software programs and online tools available for constructing phylogenetic trees. Here are some popular options:
- MEGA (Molecular Evolutionary Genetics Analysis): MEGA is a comprehensive software suite for molecular evolutionary analysis. It provides tools for sequence alignment, phylogenetic tree construction using various methods, and visualization of trees.
- PhyloSuite: PhyloSuite is a user-friendly software package that integrates multiple tools for phylogenetic analysis. It offers a range of features, including sequence alignment, tree reconstruction, model selection, and tree visualization.
- RAxML: RAxML (Randomized Axelerated Maximum Likelihood) is a fast and efficient software tool for maximum likelihood-based phylogenetic tree inference. It can handle large datasets and offers various models and options for tree construction.
- MrBayes: MrBayes is a popular software program for Bayesian inference of phylogenetic trees. It utilizes Markov chain Monte Carlo (MCMC) methods to estimate posterior probabilities of trees and model parameters.
- IQ-TREE: IQ-TREE is a fast and versatile software package for phylogenetic tree inference. It supports a wide range of models, including maximum likelihood, Bayesian inference, and ultrafast bootstrap approximation.
- PhyML: PhyML is a user-friendly software tool for maximum likelihood-based phylogenetic tree construction. It offers various substitution models, tree searching algorithms, and statistical tests.
- iTOL (Interactive Tree Of Life): iTOL is a web-based tool for interactive tree visualization and annotation. It allows users to upload their phylogenetic trees and customize the visual representation, labels, and annotations.
- Phylo.io: Phylo.io is an online platform that provides an intuitive interface for creating and exploring phylogenetic trees. It supports multiple tree formats and offers various customization options.
These are just a few examples of phylogenetic tree maker tools. The choice of software or tool depends on your specific needs, the size of your dataset, the analysis methods you prefer, and the level of expertise required.
Terminology of phylogenetic trees
- Taxon: A taxon is any group of organisms classified at a specific level, such as a species, genus, family, order, or higher taxonomic ranks.
- Node: A node, also known as a vertex, is the intersection of branches in a phylogenetic tree. It represents an ancestor from whom two or more descendant lineages diverged.
- Root: A phylogenetic tree’s root represents the most recent common ancestor of all taxa included in the tree. It is typically depicted at the tree’s base or beginning.
- Branch: A branch is an evolutionary lineage that connects a node to a tip or another node. It represents the course of evolution or diversification.
- Clade: A clade is a monophyletic group within a phylogenetic tree consisting of an ancestor and all of its descendants. It denotes a distinct evolutionary lineage and is characterized by a common ancestor.
- Tip/Leaf: A tip, also known as a leaf or terminal node, represents a taxon or group of organisms that are extant or presently living at the end of a branch. The tips of a taxonomic tree may correspond to individual species or larger taxonomic categories.
- Branch Length: The length of a branch represents the quantity of evolutionary change that has occurred along that branch. It can be measured in terms of time (for instance, millions of years) or genetic or morphological differences (for instance, DNA substitutions).
- Phylogenetic Distance: The measure of evolutionary difference or similarity between two taxa or groups. It is typically estimated using genetic or morphological data and can be quantified using a variety of techniques, such as genetic distances or character-based measures.
- Homology: Homology refers to the similarity between characteristics or traits of various organisms owing to a common ancestor. Homologous characteristics are inherited from a common ancestor and may have changed or diverged over time.
- Homoplasy: Homoplasy refers to the similarity between characteristics or traits in various organisms that is not due to a common ancestor but rather arises independently. Homoplastic characteristics may be the result of convergent evolution, parallel evolution, or evolutionary reversals.
- Bootstrap Support: Bootstrap support is a statistical confidence measure for a phylogenetic tree’s branching pattern. It is determined by repeatedly resampling the original dataset and evaluating the stability of the inferred relationships.
Limitations of phylogenetic tree
- Incomplete Data: Phylogenetic trees rely largely on available data, which may be biased or insufficient. Absence of or insufficient genetic or morphological data can result in incomplete or inaccurate trees.
- Long-Branch Attraction: Long branches on a phylogenetic tree can result in the phenomenon known as long-branch attraction. This occurs when distantly related taxa appear to be closely related due to shared evolutionary changes along their lengthy branches, leading to erroneous inferences about their relationships.
- Convergence and Parallelism: Convergent evolution, in which unrelated organisms develop similar traits as a result of similar selective pressures, can contribute to erroneous relationships in phylogenetic trees. Parallel evolution, in which closely related organisms independently acquire comparable characteristics, can also cause confusion.
- Horizontal Gene Transfer: Phylogenetic trees presume a predominantly vertical pattern of evolution, in which genetic information is transmitted from parent to offspring. However, horizontal gene transfer, the transmission of genetic material between unrelated organisms, can make the interpretation of a tree more complicated.
- Rate Variation: The variation in evolutionary rates among various lineages or genes can impede the estimation of branch lengths and the inference of relationships. Some branches may evolve more quickly or more slowly than others, creating discrepancies in the inferred relationships.
Advantages of phylogenetic tree
- Evolutionary History: Phylogenetic trees provide a graphical representation of the evolutionary history and inter-organism relationships. They aid in the comprehension of patterns of descent and divergence, disclosing the common ancestry and evolutionary history of taxa.
- Classification and Taxonomy: Modern biological classification systems are built on phylogenetic structures. They facilitate the organization and categorization of organisms according to their evolutionary relationships, allowing for a more systematic and meaningful classification of species.
- Hypothesis Testing: Phylogenetic trees allow scientists to test hypotheses concerning the evolutionary relationships and patterns of organisms. They can aid in the evaluation of competing hypotheses, the evaluation of the support for particular branches or relationships, and the direction of future investigations and research.
- Comparative Studies: Phylogenetic trees provide a structure for comparative investigations across species. By comparing the traits, behaviors, and characteristics of closely related organisms, scientists can gain a better understanding of the evolutionary patterns and processes that have shaped those traits.
- Biogeography and Conservation: Phylogenetic trees can cast light on the biogeographic distribution of organisms, allowing scientists to comprehend how species have dispersed and diversified in various regions. In addition, they can aid conservation efforts by identifying evolutionary distinct lineages or species that require special conservation measures.
Differences between a Phylogeny, an Evolutionary Tree, a Phylogenetic Tree, and a Cladogram
Phylogeny | Evolutionary Tree | Phylogenetic Tree | Cladogram | |
|---|---|---|---|---|
| Definition | Evolutionary history and relationships among organisms | Visual representation of evolutionary relationships | Graphical representation of evolutionary relationships | Diagrammatic representation of relationships based on shared characteristics |
| Scope | Broad term that encompasses various ways to represent and study evolutionary relationships | Specific type of representation of evolutionary relationships | Specific type of representation of evolutionary relationships | Specific type of representation of evolutionary relationships |
| Representation | Can be represented in various forms, such as evolutionary trees, phylogenetic trees, or cladograms | Visual representation of branching patterns and evolutionary connections | Graphical representation with branches and nodes | Diagrammatic representation using branches and shared derived traits |
| Methodology | Can be constructed using various methods and data types | Can be constructed using various methods and data types | Primarily constructed using distance-based, parsimony, maximum likelihood, or Bayesian methods | Primarily constructed using shared derived traits |
| Information | Represents the overall evolutionary history and relationships | Represents the overall evolutionary history and relationships | Represents the evolutionary relationships and often includes branch lengths | Represents relationships based on shared derived traits, often without branch lengths |
| Branch Lengths | May or may not include branch lengths, depending on the representation | May or may not include branch lengths, depending on the representation | Often includes branch lengths, representing genetic or evolutionary distances | Does not typically include branch lengths |
| Representation of Character Evolution | Can incorporate information about character evolution, such as ancestral states and character changes | Can incorporate information about character evolution, such as ancestral states and character changes | Can incorporate information about character evolution, such as ancestral states and character changes | Focuses on relationships based on shared derived traits rather than character evolution |
Phylogenetic tree vs cladogram
| Criteria | Phylogenetic Tree | Cladogram |
|---|---|---|
| Representation | Represents evolutionary relationships, including branching patterns, common ancestry, and divergence times (if available). | Represents only the branching patterns of relationships, without indicating branch lengths or time scales. |
| Branch Lengths | Branch lengths can represent various factors such as genetic distance, time, or evolutionary change. Longer branches may indicate more divergence or greater genetic differences. | Branch lengths are typically not included. All branches are assumed to be of equal length, indicating only the order of branching events. |
| Time Information | Can include explicit time scales, indicating estimated time of divergence between lineages. | Does not include explicit time information. |
| Taxonomic Hierarchy | Can represent hierarchical taxonomic relationships, indicating different taxonomic ranks such as families, genera, or species. | Does not directly represent taxonomic hierarchy. |
| Evolutionary Patterns | Provides insights into the patterns and processes of evolutionary change, including the relationships between different lineages, the timing of evolutionary events, and the rate of molecular evolution. | Focuses primarily on identifying shared derived characteristics (synapomorphies) among groups to define clades. |
| Representation of Relationships | Shows the degree of relatedness between different groups through the length and branching angles of the branches. | Shows the presence or absence of shared derived characteristics to define clades, but does not indicate the degree of relatedness or evolutionary distance between different groups. |
| Example | A phylogenetic tree might show the evolutionary relationships between different primate species, indicating when they branched off from a common ancestor and the estimated divergence times. | A cladogram might show the branching pattern of reptiles, indicating the relationships between different reptile groups based on shared derived characteristics such as scales and shelled eggs. |
Phylogenetic Tree Example
1. Animal Phylogenetic Tree
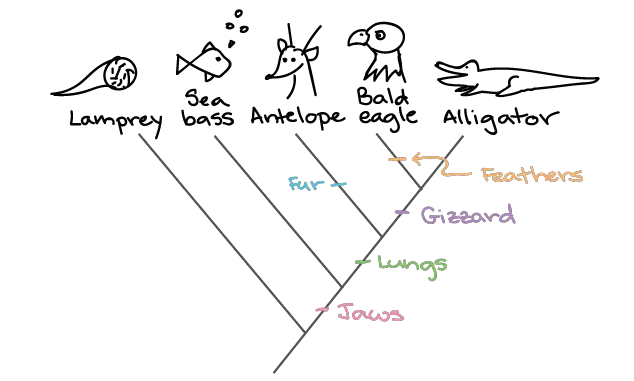
- This tree shows the evolutionary relationships among major animal groups.
- It depicts the common ancestry of animals and the branching patterns leading to the diversification of different animal phyla.
- The tree reveals that animals evolved from a common ancestor and branched into different lineages, including chordates, arthropods, mollusks, and others.
2. Plant Phylogenetic Tree
- This tree illustrates the evolutionary relationships among major plant groups.
- It showcases the divergence of plant lineages from a common ancestor and the subsequent evolution of mosses, ferns, gymnosperms, and angiosperms (flowering plants).
- The tree highlights the relationships between different plant groups based on shared derived traits and genetic information.

3. Bacterial Phylogenetic Tree
- This tree represents the evolutionary relationships among various bacterial taxa.
- It demonstrates the diversification of bacteria into different phyla, classes, orders, and families.
- The tree reflects the evolutionary history of bacteria, highlighting the relationships between different groups based on genetic and phenotypic characteristics.
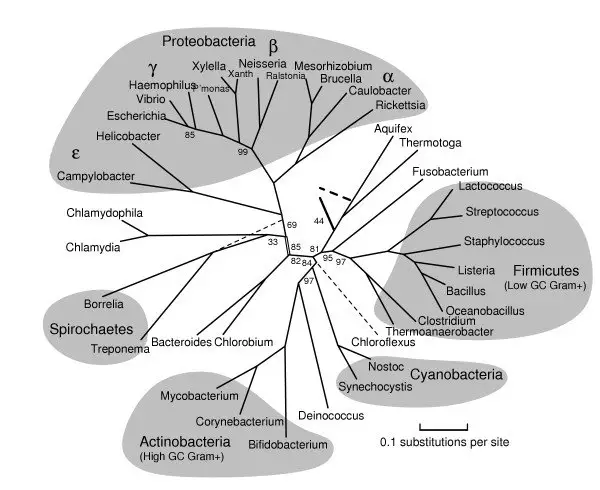
4. Fungal Phylogenetic Tree
- This tree showcases the evolutionary relationships among major fungal groups.
- It demonstrates the divergence of fungal lineages from a common ancestor and the subsequent evolution of different fungal phyla, including Ascomycota, Basidiomycota, Zygomycota, and others.
- The tree reveals the relationships between different fungal groups based on shared ancestry and genetic information.
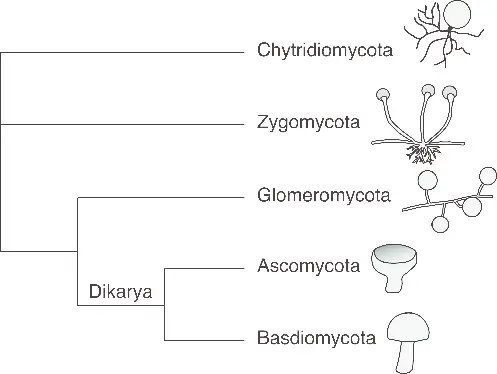
5. Human phylogenetic tree
- Humans, several non-human primates, and four classic model organisms used in aging research are depicted on a phylogenetic tree.
- The horizontal axis represents the number of millions of years since the divergence of two species. The divergence between humans and marmosets is estimated to have occurred approximately 42,6 million years ago. The divergence between humans and fruit flies occurred roughly 782 million years ago.
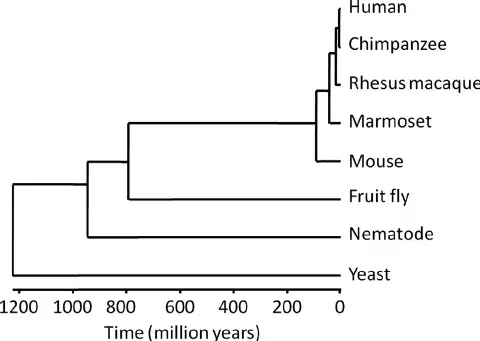
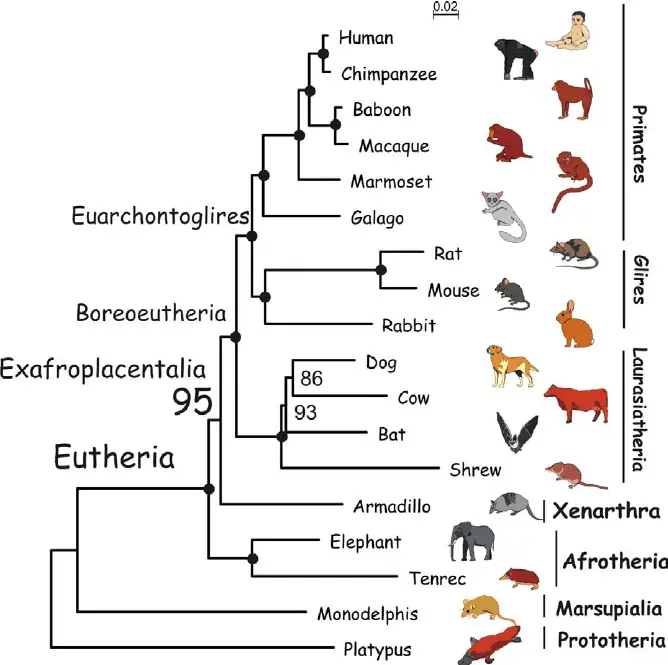
6. Primate phylogenetic tree
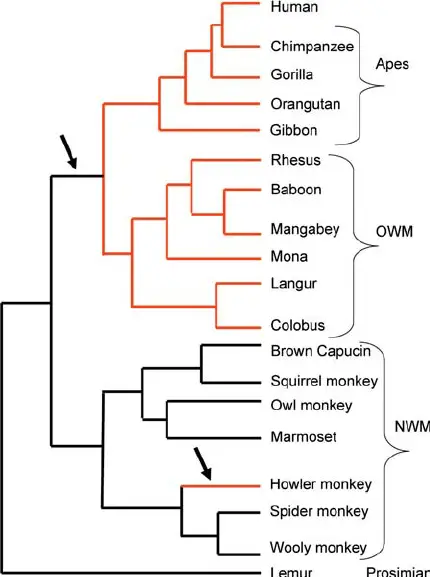
7. Vertebrate phylogenetic tree
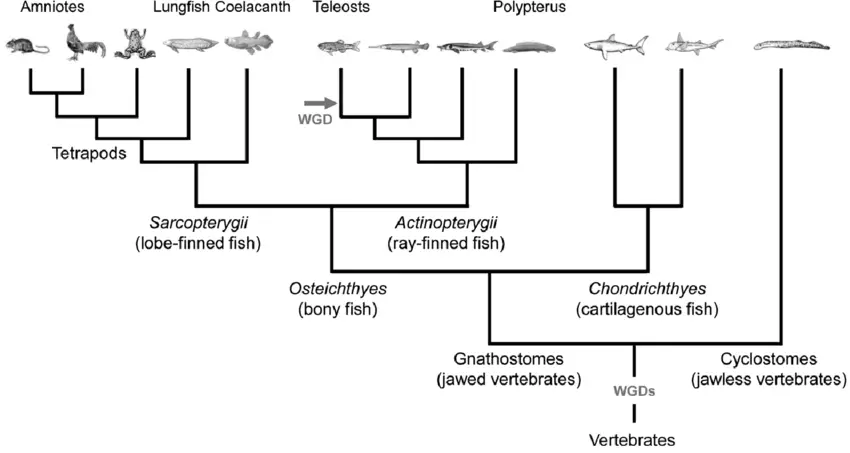
8. Phylogenetic tree mammals
Phylogenetic tree practice
- Which evolutionary concept is represented by the branch points in a phylogenetic tree?
- How are the lengths of branches in a phylogenetic tree typically interpreted?
- What is the purpose of using outgroup taxa in phylogenetic tree construction?
- How does the use of different algorithms (e.g., maximum likelihood, maximum parsimony) affect the construction of phylogenetic trees?
- What is the difference between a polytomy and a resolved branch in a phylogenetic tree?
- How can the bootstrap value or posterior probability be used to assess the support for a specific branch in a phylogenetic tree?
- Why is it important to consider multiple lines of evidence, such as molecular data and morphological traits, when constructing a phylogenetic tree?
- Explain the concept of convergent evolution and how it can influence the interpretation of phylogenetic trees.
- How can phylogenetic trees help in understanding the origins and evolutionary relationships of specific traits or adaptations?
- Discuss the challenges and limitations of inferring phylogenetic relationships when dealing with incomplete or ambiguous data.
FAQ
What is a phylogenetic tree?
A phylogenetic tree is a diagrammatic representation of the evolutionary relationships among different organisms or sequences. It illustrates the branching patterns and common ancestry of species or genes.
How are phylogenetic trees constructed?
Phylogenetic trees are constructed using various methods, such as maximum likelihood, maximum parsimony, or distance-based algorithms. These methods analyze genetic or morphological data to infer evolutionary relationships.
What is the purpose of a phylogenetic tree?
Phylogenetic trees help us understand the evolutionary history, relatedness, and diversification of organisms. They provide insights into the patterns of evolutionary change and can be used to study various biological phenomena.
What do the branches and nodes represent in a phylogenetic tree?
The branches in a phylogenetic tree represent evolutionary relationships and show the divergence of lineages. Nodes, or branch points, represent common ancestors from which different lineages have evolved.
How are branch lengths interpreted in a phylogenetic tree?
Branch lengths in a phylogenetic tree can represent different factors, such as genetic distance, time, or evolutionary change. Longer branches typically indicate more divergence or greater genetic differences between taxa.
What is the difference between a cladogram and a phylogenetic tree?
A cladogram is a type of phylogenetic tree that represents only the branching pattern of relationships, without indicating branch lengths or time scales. A phylogenetic tree includes additional information such as branch lengths and can provide more detailed insights into evolutionary history.
How can I interpret the support values in a phylogenetic tree?
Support values, such as bootstrap values or posterior probabilities, indicate the statistical support for specific branches or nodes in a phylogenetic tree. Higher support values suggest more confidence in the inferred relationships.
Can phylogenetic trees be used to study human evolution?
Yes, phylogenetic trees are commonly used to study human evolution. They help trace the evolutionary relationships among different hominid species and shed light on the origins and diversification of our lineage.
What are the limitations of phylogenetic trees?
Phylogenetic trees are based on available data and assumptions, so they can be affected by data quality, incomplete sampling, or inaccuracies in the underlying models. Additionally, the interpretation of tree topologies can sometimes be challenging due to complex evolutionary processes.
How can phylogenetic trees inform conservation biology?
Phylogenetic trees can help identify evolutionary distinct species, prioritize conservation efforts based on genetic diversity, and understand the evolutionary origins and relationships of endangered or threatened species.
Which type of evidence is least likely to result in changes to a phylogenetic tree?
The type of evidence that is least likely to result in changes to a phylogenetic tree is strong and well-supported evidence from multiple independent sources. When multiple lines of evidence consistently support a particular phylogenetic tree topology, it becomes more robust and less likely to change.
For example, if different types of data, such as molecular sequences, morphological characteristics, and fossil records, all consistently support the same phylogenetic relationships, it provides a strong foundation for the tree’s accuracy. In such cases, it becomes less likely that new evidence or analysis will drastically alter the overall tree topology.
On the other hand, if the evidence is weak, conflicting, or based on a single source, there is a higher probability of changes to the phylogenetic tree as new data becomes available or alternative analytical methods are applied. In such cases, further research, increased sampling, or the incorporation of additional data can lead to revisions or refinements in the phylogenetic relationships.
References
- https://www.khanacademy.org/science/ap-biology/natural-selection/phylogeny/a/phylogenetic-trees#:~:text=A%20phylogenetic%20tree%20is%20a,a%20series%20of%20common%20ancestors.
- https://en.wikipedia.org/wiki/Phylogenetic_tree
- https://bioprinciples.biosci.gatech.edu/module-1-evolution/phylogenetic-trees/
- https://www.geeksforgeeks.org/phylogenetic-tree/
- https://courses.lumenlearning.com/wm-biology1/chapter/reading-phylogenetic-trees-2/


