Table of Contents
What is Polymerase chain reaction (PCR)?
- Polymerase chain reaction (PCR) is a widely used enzymatic process that allows scientists to replicate a specific region of DNA, resulting in the production of many copies of a particular DNA sequence. PCR combines the principles of complementary nucleic acid hybridization and nucleic acid replication to amplify a single copy of a target DNA sequence, even if it is undetectable by standard hybridization methods. This amplification process can generate millions to billions of copies of the target DNA in a relatively short period of time, providing an abundant amount of DNA for further analysis.
- PCR was invented in 1983 by American biochemist Kary Mullis at Cetus Corporation, for which he was awarded the Nobel Prize in Chemistry in 1993, jointly with biochemist Michael Smith. Since its development, PCR has become a fundamental technique in genetic testing and research. It is used in a wide range of applications, including the analysis of ancient DNA samples, identification of infectious agents, gene cloning and manipulation, DNA sequencing, diagnosis of genetic disorders, forensic science, and detection of pathogens in infectious diseases.
- The PCR method typically relies on thermal cycling, which involves exposing the reaction to repeated cycles of heating and cooling. These temperature changes facilitate specific reactions, such as DNA melting and enzyme-driven DNA replication. Two key components of PCR are primers and a DNA polymerase. Primers are short single-stranded DNA fragments, known as oligonucleotides, that are complementary to the target DNA region. The DNA polymerase is the enzyme responsible for assembling new DNA strands using the primers and free nucleotides.
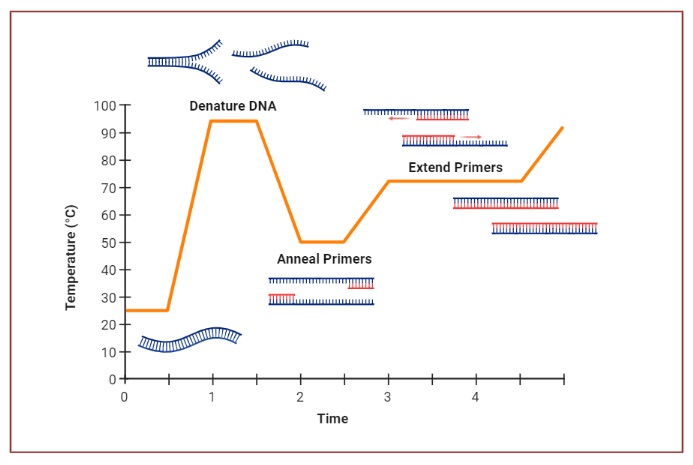
- The PCR process involves several steps. In the initial denaturation step, the double-stranded DNA template is heated to a high temperature, causing the two strands to separate. Then, the temperature is lowered, allowing the primers to bind to the complementary sequences of the DNA. Once the primers are bound, the DNA polymerase synthesizes a new DNA strand using the template DNA and free nucleotides. This process is repeated multiple times, with each cycle resulting in the amplification of the target DNA. The DNA generated in one cycle becomes the template for the next, leading to an exponential increase in the amount of DNA.
- To ensure the success of PCR, a heat-stable DNA polymerase is typically used, such as Taq polymerase, which was originally isolated from the bacterium Thermus aquaticus. Taq polymerase can withstand the high temperatures of the denaturation step without denaturing itself. Prior to the use of Taq polymerase, DNA polymerase had to be manually added after each cycle, which was a time-consuming and expensive process.
- PCR has revolutionized various fields of research and diagnostics. It has enabled scientists to amplify and study minute amounts of DNA, providing valuable insights into genetic information and facilitating advancements in fields such as medicine, forensics, and evolutionary biology.
Definition of Polymerase chain reaction (PCR)
Polymerase chain reaction (PCR) is a widely used enzymatic process that rapidly and exponentially amplifies a specific region of DNA, producing millions to billions of copies of a particular DNA sequence.
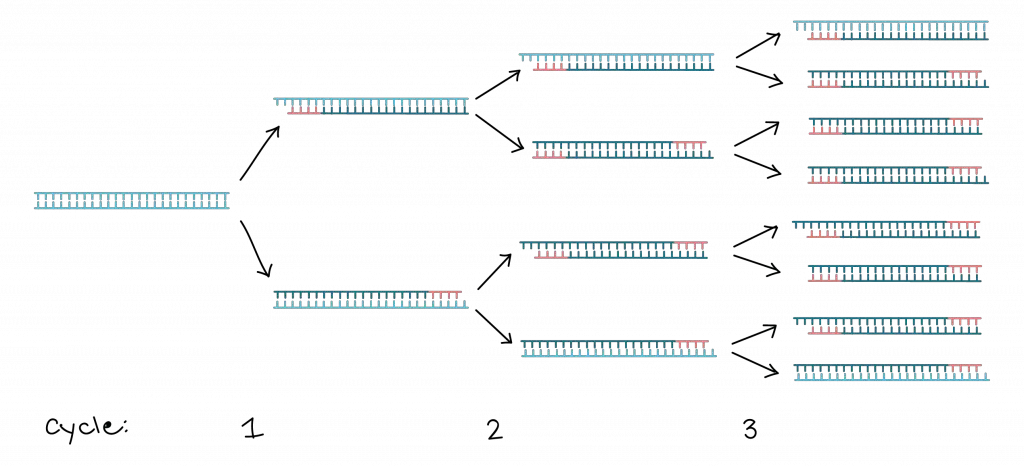
Principle of Polymerase chain reaction
The principle of polymerase chain reaction (PCR) involves the amplification of a specific segment of DNA through a series of temperature cycles. The process begins with denaturation, where the target DNA is heated to separate the two strands, resulting in single-stranded DNA. Next, specific primers, designed to bind to each target DNA strand, are added. These primers serve as starting points for DNA synthesis.
During the primer annealing step, the reaction mixture is cooled to a temperature that allows the primers to bind to their complementary sequences on the target DNA. Once the primers are bound, DNA polymerase, an enzyme that synthesizes new DNA strands, extends the primers by adding complementary nucleotides. This primer extension step occurs at a temperature optimal for DNA polymerase activity.
The cycling steps in PCR involve repeating the denaturation, primer annealing, and primer extension steps multiple times. Each cycle results in the duplication of the target DNA. After 25 to 30 cycles, the number of copies of the target DNA can reach at least 107 due to the exponential amplification.
PCR typically utilizes a series of temperature changes, known as thermal cycling, to facilitate the different steps. The cycling steps usually consist of three temperature shifts: denaturation, primer annealing, and primer extension. Denaturation is typically performed at a high temperature (around 94-96°C) to separate the DNA strands. During primer annealing, the temperature is lowered (around 45-60°C) to allow the primers to bind to their target sequences. Finally, primer extension occurs at a temperature optimal for DNA polymerase activity, usually around 72°C.
PCR methods often include multiple cycles of these temperature changes, starting and ending with a hold step at higher and lower temperatures, respectively. The hold steps ensure the completion of product extension and allow for the analysis or storage of the final PCR product. While most PCR methods amplify DNA fragments up to around 10 kilo base pairs (kb), some techniques can amplify even larger fragments, reaching up to 40 kb.
Requirements for PCR
Requirements for PCR involve several components and conditions that are necessary for the successful amplification of DNA. Here are the key requirements for PCR:
- DNA template: The DNA template is the target sequence that will be amplified. It can be isolated from various sources, such as blood, tissue, forensics specimens, or microbial cells. The DNA template should contain the target sequence that needs to be amplified.
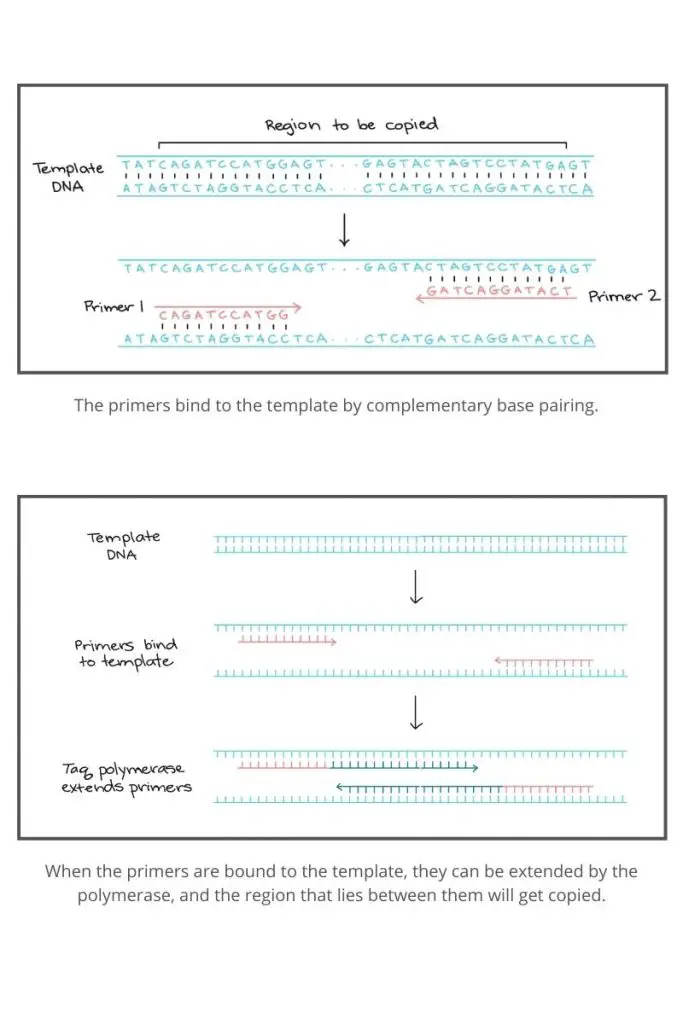
- Primers: Primers are short, single-stranded DNA molecules that bind to the DNA template and provide a starting point for DNA synthesis. Two primers are used in PCR: the forward primer, which binds to one strand of the target DNA, and the reverse primer, which binds to the complementary strand. Primers need to be designed specifically for the target DNA sequence to ensure specificity and efficiency of amplification.
- DNA polymerase: DNA polymerase is an enzyme that synthesizes new DNA strands by adding nucleotides to the primer. The most commonly used DNA polymerase in PCR is Taq DNA polymerase, derived from the bacterium Thermus aquaticus. Taq polymerase is thermostable and can withstand the high temperatures used during denaturation. Other DNA polymerases with proofreading capabilities, such as Pfu or KOD polymerase, can also be used to minimize errors during amplification.
- Nucleotides: PCR requires all four deoxynucleoside triphosphates (dNTPs) – adenine (A), guanine (G), cytosine (C), and thymine (T) – to synthesize new DNA strands. These nucleotides are added to the reaction mixture at equal concentrations.
- Buffer solution: The PCR buffer provides the necessary conditions and chemicals for optimal activity and stability of the DNA polymerase. It typically contains Tris-HCl to maintain the pH, KCl to stabilize primer-template annealing, and MgCl2 as a cofactor for DNA polymerase activity. Other additives may be included depending on the specific PCR protocol.
- Monovalent and divalent cations: Potassium chloride (KCl) is commonly used as a monovalent cation in PCR reactions to optimize the conditions for DNA amplification. Divalent cations, such as magnesium (Mg2+), are essential as cofactors for DNA polymerase activity. Mg2+ ions are typically included in the PCR buffer, although manganese (Mn2+) can be used in certain cases, such as DNA mutagenesis.
- PCR tube: PCR is performed in small, thin-walled plastic tubes called PCR tubes. These tubes allow for efficient thermal conductivity and temperature equilibration during the thermal cycling process.
- Thermal cycler: A thermal cycler, also known as a thermocycler, is a device used to rapidly heat and cool the PCR reaction mixture during the temperature cycling steps. Modern thermocyclers use the Peltier effect to achieve precise temperature control. They are also equipped with heated lids to prevent condensation and evaporation of the reaction mixture.
These requirements, when combined and optimized, enable the efficient amplification of specific DNA sequences using PCR.
Preparation of PCR Reaction Mixture
Add these following ingredients to a PCR tube;
- Sterile Water
- 10X Assay Buffer
- 10 mM dNTP Mix
- Template DNA (100 ng/µl)
- Forward Primer (100 ng/µl)
- Reverse Primer (100 ng/µl)
- Taq DNA Polymerase (3 U/µl)
Note: Tap the tube for 1–2 seconds to mix the contents thoroughly. Then, add 25 μl of mineral oil in the tube to avoid evaporation of the contents. Place the tube in the thermocycler block and set the program to get DNA amplification.
Steps of Polymerase Chain Reaction (PCR)
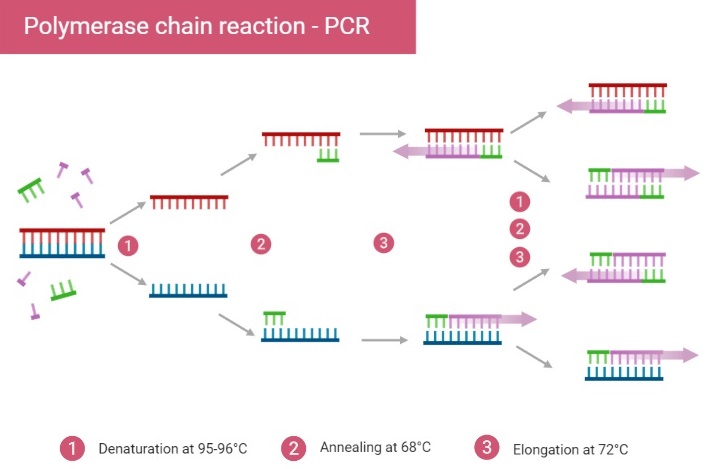
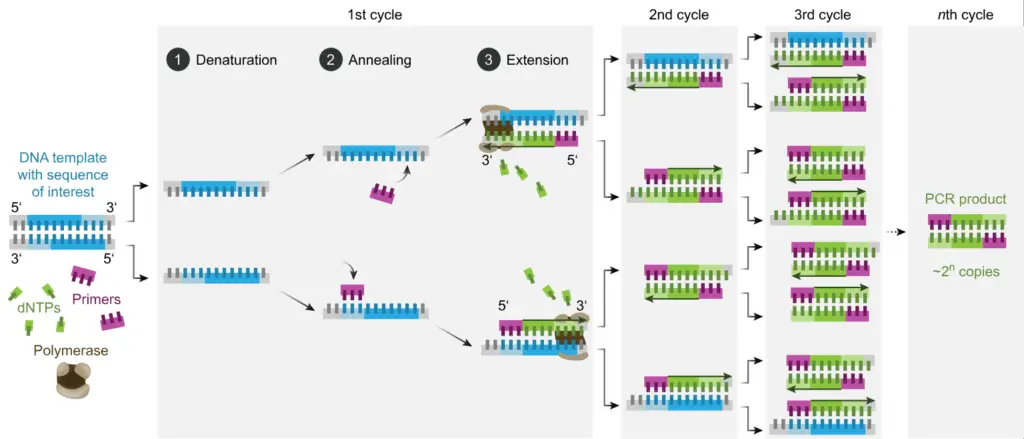
The polymerase chain reaction (PCR) is a powerful technique used to amplify specific DNA sequences. It involves a series of temperature changes known as thermal cycles. The steps involved in PCR are as follows:
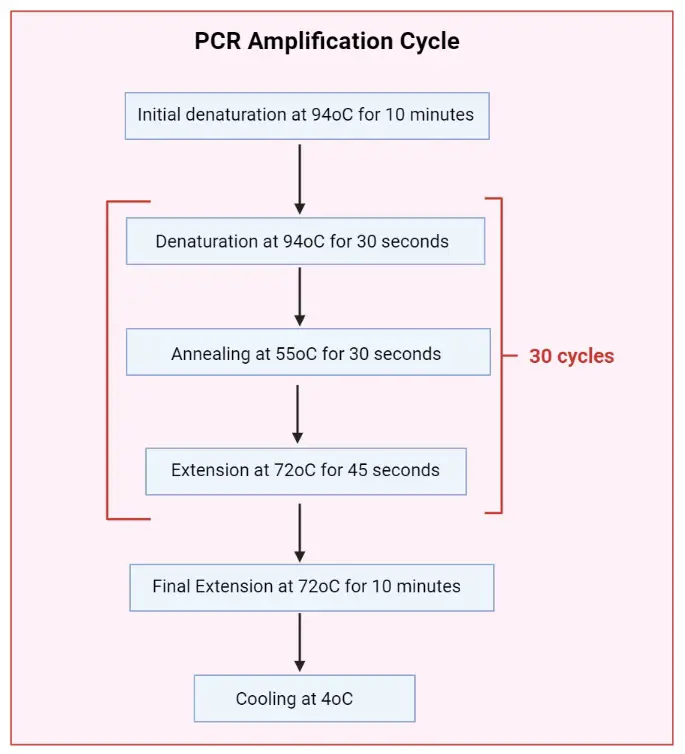
- Initialization: This step is required for DNA polymerases that need heat activation. The reaction chamber is heated to a temperature of 94-96 °C (or 98 °C for extremely thermostable polymerases) and held for 1-10 minutes.
- Denaturation: The reaction chamber is heated to 94-98 °C for 20-30 seconds. This causes the double-stranded DNA template to denature, separating it into two single-stranded DNA molecules.
- Annealing: The temperature is lowered to 50-65 °C for 20-40 seconds. During this step, short DNA sequences called primers anneal to each of the single-stranded DNA templates. Primers are designed to bind to specific regions flanking the target DNA sequence.
- Extension/Elongation: The temperature is raised to the optimal activity temperature of the DNA polymerase used, typically around 72-80 °C. The DNA polymerase synthesizes a new DNA strand complementary to the template strand by adding nucleotides (dNTPs) in the 5′-to-3′ direction. This step extends the primers and synthesizes new DNA strands.
- Cycling: Steps 2 to 4 (denaturation, annealing, and extension) are repeated for 20-40 cycles. With each cycle, the target DNA is doubled, resulting in exponential amplification of the specific DNA sequence.
- Final elongation: After the last PCR cycle, a final elongation step is performed at a temperature of 70-74 °C for 5-15 minutes to ensure complete elongation of any remaining single-stranded DNA.
- Final hold: The reaction chamber is cooled to 4-15 °C for an indefinite time. This step allows for short-term storage of the PCR products.
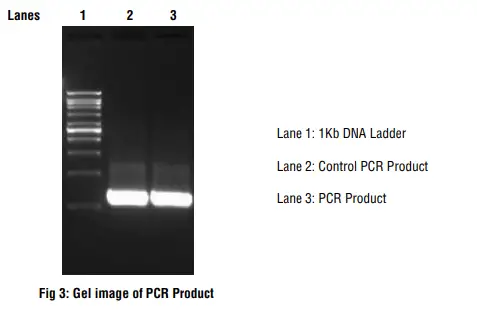
To verify the success of the PCR, agarose gel electrophoresis can be used. The amplified DNA products are separated based on size using a DNA ladder as a reference.
PCR is a versatile technique used in various applications, such as gene expression analysis, genetic testing, forensics, and molecular biology research. Its ability to amplify specific DNA sequences quickly and efficiently has revolutionized many fields of study.
Description of Each Steps of Polymerase Chain Reaction (PCR)
1. Initialization
- Initialization is an important step in the polymerase chain reaction (PCR) process, particularly for DNA polymerases that require heat activation through hot-start PCR. This step is performed at the beginning of the PCR reaction and involves heating the reaction chamber to a specific temperature.
- The purpose of the initialization step is to ensure that the DNA polymerase enzyme is fully activated and ready to perform its function of synthesizing new DNA strands. Not all DNA polymerases require this heat activation, but for those that do, initialization becomes necessary.
- During initialization, the reaction chamber is typically heated to a temperature range of 94-96 °C (201-205 °F). However, if extremely thermostable polymerases are used, the temperature can be elevated to 98 °C (208 °F). The specific temperature used depends on the requirements of the particular DNA polymerase being employed.
- The duration of the initialization step can vary and is generally held for 1-10 minutes. This time allows the DNA polymerase to undergo the necessary conformational changes, enabling it to reach its optimal state for DNA synthesis.
- By subjecting the reaction chamber to this elevated temperature during initialization, any residual enzyme activity that could lead to non-specific amplification or primer-dimer formation is minimized or eliminated. This hot-start approach helps improve the specificity and efficiency of the PCR reaction by preventing undesired DNA amplification before the actual cycling stages begin.
- Overall, the initialization step in PCR is crucial for ensuring the activation of DNA polymerases that require heat activation. By heating the reaction chamber to a specific temperature and holding it for a designated period, the DNA polymerase enzyme is prepared for subsequent steps, such as denaturation, annealing, and extension, leading to successful DNA amplification.
2. Denaturation
- Denaturation is a crucial step in the polymerase chain reaction (PCR) process. It is the first regular cycling event that occurs after the initialization step. Denaturation involves heating the reaction chamber to a temperature range of 94-98 °C (201-208 °F) for a duration of 20-30 seconds.
- The primary objective of the denaturation step is to induce the melting or separation of the double-stranded DNA template into two single-stranded DNA molecules. This process is achieved by breaking the hydrogen bonds that hold the complementary bases of the DNA strands together. As a result, the double-stranded DNA template is transformed into single-stranded DNA templates, which serve as the starting point for subsequent stages of the PCR reaction.
- The high temperature used during denaturation disrupts the hydrogen bonding between the two complementary DNA strands. The DNA molecule unwinds, and the two strands separate from each other. The heat provides the energy required to overcome the hydrogen bonding forces between the base pairs, thereby promoting the separation of the strands. This denaturation step is crucial because it allows access to the DNA template for the binding of primers in the following annealing step.
- By generating single-stranded DNA templates, denaturation enables the primers to anneal to their complementary sequences during the next stage of PCR. The denaturation step ensures that the DNA template is in a suitable conformation for efficient primer binding and subsequent DNA synthesis.
- The duration of the denaturation step is relatively short, typically ranging from 20 to 30 seconds. This short exposure to high temperature is sufficient to disrupt the hydrogen bonds and separate the DNA strands without causing significant damage to the DNA molecules.
- In summary, denaturation is a critical step in the PCR process. By subjecting the reaction chamber to a high temperature for a short period, the double-stranded DNA template is melted into single-stranded DNA templates, allowing for efficient primer binding and subsequent amplification of the target DNA sequence.
3. Annealing
- Annealing is a crucial step in the polymerase chain reaction (PCR) process that follows denaturation. During annealing, the reaction temperature is lowered to a range of 50-65 °C (122-149 °F) for a duration of 20-40 seconds. This temperature range allows for the specific binding of primers to each of the single-stranded DNA templates generated during denaturation.
- In PCR, two different primers are typically included in the reaction mixture. Each primer is designed to complement a short sequence of nucleotides at the 3′ end of one of the single-stranded DNA templates containing the target region. The primers themselves are single-stranded sequences and are much shorter than the overall length of the target region.
- The annealing temperature is a critical parameter to determine as it directly influences the efficiency and specificity of primer binding. The temperature must be low enough to enable the hybridization or binding of the primers to their complementary sequences on the single-stranded DNA templates. However, it must also be high enough to ensure specific binding, where the primers bind only to their perfectly complementary sequences and nowhere else.
- If the annealing temperature is too low, the primers may bind imperfectly to non-specific regions of the DNA template, leading to nonspecific amplification and undesirable products. On the other hand, if the temperature is too high, the primers may fail to bind at all, resulting in no amplification.
- A typical annealing temperature is set to be around 3-5 °C below the melting temperature (Tm) of the primers used. The Tm represents the temperature at which stable hydrogen bonds are formed between complementary bases. During annealing, the primer sequence should closely match the template sequence to allow for stable and specific binding.
- During the annealing step, the DNA polymerase enzyme binds to the primer-template hybrid formed by the annealing of primers to their complementary sequences on the single-stranded DNA templates. The polymerase, once bound, initiates the synthesis of a new DNA strand complementary to the DNA template. This DNA synthesis marks the beginning of DNA amplification in PCR.
- In summary, annealing is a critical step in PCR where the reaction temperature is lowered to enable the specific binding of primers to their target sequences on the single-stranded DNA templates. The proper determination of the annealing temperature ensures efficient and specific primer binding, facilitating subsequent DNA synthesis by the polymerase enzyme.
4. Extension/elongation
- The extension/elongation step is a crucial part of the polymerase chain reaction (PCR) process, following denaturation and annealing. The temperature for this step depends on the DNA polymerase being used. For example, the thermostable DNA polymerase Taq polymerase has an optimum activity temperature of approximately 75–80 °C (167–176 °F), although a temperature of 72 °C (162 °F) is commonly used with this enzyme.
- During the extension/elongation step, the DNA polymerase synthesizes a new DNA strand that is complementary to the DNA template strand. This synthesis occurs by adding free deoxyribonucleotides (dNTPs) from the reaction mixture. The dNTPs are complementary to the template sequence and are added in the 5′-to-3′ direction, which is the direction of DNA synthesis. The polymerase enzyme catalyzes the condensation reaction, joining the 5′-phosphate group of the dNTPs with the 3′-hydroxy group at the end of the nascent DNA strand. This process results in the elongation of the DNA strand.
- The time required for elongation depends on factors such as the specific DNA polymerase being used and the length of the DNA target region being amplified. As a general guideline, most DNA polymerases can polymerize approximately a thousand bases per minute at their optimal temperature.
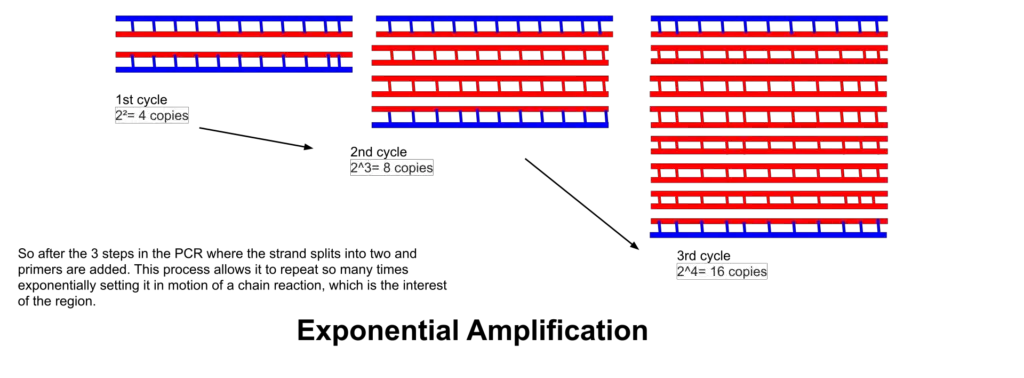
- Under optimal conditions with no limitations due to limiting substrates or reagents, each extension/elongation step in PCR doubles the number of DNA target sequences. This is because the original template strands, as well as all newly generated strands, become template strands for the next round of elongation. This phenomenon leads to exponential (geometric) amplification of the specific DNA target region as PCR cycles progress.
- The processes of denaturation, annealing, and elongation together constitute a single cycle in PCR. However, multiple cycles are necessary to amplify the DNA target to millions of copies. The formula used to calculate the number of DNA copies formed after a given number of cycles is 2^n, where n represents the number of cycles. For example, a reaction set for 30 cycles would result in 2^30, or 1,073,741,824 copies of the original double-stranded DNA target region.
- In summary, the extension/elongation step of PCR involves the DNA polymerase synthesizing a new DNA strand complementary to the DNA template strand by adding dNTPs. Each elongation step doubles the number of DNA target sequences, leading to exponential amplification. Multiple cycles are performed to achieve the desired level of DNA amplification.
5. Final elongation
- The final elongation step in the polymerase chain reaction (PCR) is an optional but often employed step that occurs after the last PCR cycle. Its purpose is to ensure that any remaining single-stranded DNA in the reaction mixture is fully elongated, leading to the completion of DNA synthesis.
- During the final elongation step, the reaction temperature is typically set to a range of 70–74 °C (158–165 °F). This temperature range is chosen because it supports the optimal activity of most polymerases used in PCR. By maintaining this temperature for a duration of 5–15 minutes, any remaining single-stranded DNA templates present in the reaction mixture can be effectively elongated.
- The rationale behind this step is to allow additional time for the DNA polymerase to complete the synthesis of any partially elongated DNA strands. By prolonging the incubation period, any incomplete extensions or gaps in the synthesized DNA strands can be filled, resulting in more complete and fully elongated double-stranded DNA molecules.
- While the final elongation step is not strictly required for successful PCR amplification, it can be beneficial in certain applications. It ensures that all available single-stranded DNA templates are thoroughly extended, reducing the chances of incomplete or truncated amplification products. Additionally, it may be particularly useful when the amplified DNA will be used for downstream applications that require high-fidelity DNA synthesis.
- In summary, the final elongation step in PCR is an optional step performed at a temperature of 70–74 °C (158–165 °F) for 5–15 minutes after the last PCR cycle. It allows for the completion of DNA synthesis by ensuring that any remaining single-stranded DNA templates are fully elongated. Although not always necessary, this step can contribute to improved amplification results and more reliable downstream applications.
6. Final hold
- The final step in the polymerase chain reaction (PCR) is the “final hold.” It involves cooling the reaction chamber to a temperature range of 4–15 °C (39–59 °F) and maintaining this temperature for an indefinite period of time. This step is typically performed after the completion of the PCR amplification cycles.
- The purpose of the final hold is to provide a suitable environment for short-term storage of the PCR products. By reducing the temperature to a cool range, it helps to stabilize and preserve the amplified DNA molecules until further analysis or processing can be carried out. The specific temperature within the range of 4–15 °C may vary depending on the specific requirements of the experiment or protocol.
- During the final hold, the reaction is essentially paused, and the PCR products are held at a low temperature. This allows for convenient handling and storage of the amplified DNA without degradation or loss of integrity. The duration of the final hold can vary depending on the immediate needs of the experiment or the time required for subsequent steps.
- The final hold temperature range of 4–15 °C is commonly achieved using a thermal cycler or a refrigerated incubation chamber. The duration of the hold can be several minutes to several hours or even overnight, depending on the experimental design and the stability requirements of the PCR products.
- By employing the final hold, researchers can temporarily store the PCR products before further analysis, such as gel electrophoresis, sequencing, or other downstream applications. This step helps to maintain the integrity of the amplified DNA and allows flexibility in the timing of subsequent experiments or procedures.
- In summary, the final hold in PCR involves cooling the reaction chamber to a temperature range of 4–15 °C for an indefinite period of time. It serves as a short-term storage step, providing a stable environment for the PCR products until they can be further analyzed or processed. This final hold temperature range allows for the preservation of the amplified DNA molecules, enabling convenient handling and flexibility in experimental workflows.
Stages of PCR
The polymerase chain reaction (PCR) can be divided into three stages based on the progress of the reaction: exponential amplification, leveling off, and plateau.
- Exponential amplification: In this stage, the PCR reaction undergoes exponential growth, leading to a rapid increase in the amount of the desired DNA product. Each cycle of PCR doubles the amount of DNA, assuming 100% reaction efficiency. For example, after 30 cycles, a single copy of DNA can be amplified to up to 1,000,000,000 (one billion) copies. This stage demonstrates the remarkable amplification power of PCR, allowing the replication of a specific DNA sequence in a controlled laboratory setting. Even minute quantities of the target DNA are sufficient for detection and analysis.
- Leveling off stage: As the PCR reaction progresses, the amplification rate starts to slow down. This stage is characterized by a decrease in the rate of product accumulation. Several factors contribute to this slowdown. The DNA polymerase enzyme may begin to lose its activity over time, reducing its efficiency in synthesizing new DNA strands. Additionally, the consumption of reagents, such as deoxynucleotide triphosphates (dNTPs) and primers, gradually depletes their availability, limiting their contribution to the reaction. These factors collectively lead to a reduced amplification rate compared to the earlier exponential phase.
- Plateau: In the plateau stage, no further increase in product accumulation is observed. This occurs when the reagents and enzyme required for the PCR reaction become completely exhausted. As the reaction progresses and continues through multiple cycles, the limited availability of dNTPs and primers prevents further DNA synthesis. Additionally, the DNA polymerase enzyme may become fully utilized or degraded, further hindering the production of new DNA strands. The reaction reaches a point where no significant amplification occurs, and the amount of product levels off.
These three stages of PCR demonstrate the dynamic nature of the reaction and highlight the importance of optimizing reaction conditions, including reagent concentrations and cycling parameters, to achieve maximum amplification efficiency. The exponential amplification stage showcases the powerful capability of PCR to rapidly generate millions or billions of copies of a specific DNA target. However, it is crucial to monitor the reaction progress and consider the limitations imposed by reagent availability and enzyme activity as the reaction progresses into the leveling off and plateau stages.
Gel electrophoresis to visualize the results of PCR
After completion of the PCR, perform agarose gel electrophoresis. Compare the amplified product with the ladder and determine its size.
Magnitude of amplification
- PCR (Polymerase Chain Reaction) is a powerful technique that allows for the exponential amplification of target DNA templates through multiple cycling steps. The magnitude of amplification achieved in PCR can be calculated using a simple formula: “2^n,” where “n” represents the number of cycles performed [27, 28]. This formula indicates that the number of DNA copies produced doubles with each cycle.
- For example, let’s consider a PCR set with 36 cycles. Applying the formula “2^36,” we find that this PCR would result in approximately 68 billion copies of the target DNA template. This demonstrates the remarkable amplification potential of PCR.
- Under optimal conditions and with high efficiency, a PCR reaction performed in a 50 ml volume can generate substantial amounts of amplified DNA. Even with minimal efficiency, after 35-40 cycles, this PCR could produce approximately 0.2 mg of a 150 bp DNA fragment from only 100 template molecules. The molar weight of the fragment is typically around 99,000 Da.
- These calculations illustrate how PCR can generate an enormous number of DNA copies from a minute starting amount. The exponential amplification achieved through successive cycles allows for the detection, analysis, and study of DNA targets that may be present in low abundance. PCR’s ability to generate such a high magnitude of amplification has made it an indispensable tool in various scientific and diagnostic applications.
Validating PCR
Validating PCR is an essential step to ensure the success and accuracy of the amplification process. Several methods can be employed to validate PCR results. Let’s explore some of the common validation techniques:
- Ethidium Bromide Staining: Ethidium bromide (EtBr) is a commonly used dye for the staining of amplified DNA products. It intercalates between the base pairs of the double helix and can be visualized under UV light. EtBr has UV absorbance maxima at 300 and 360 nm, with an emission maximum at 590 nm. By staining the DNA bands, the presence and intensity of the amplified product can be determined. The detection limit for DNA bound to ethidium bromide is typically around 0.5-5.0 ng per band.
- Three Primer Combination Approach: In some cases, a three primer combination approach can be employed for a more cost-effective end-labeling of PCR products. This approach involves using a fluorescently labeled universal primer, modified locus-specific primers, and 5′ universal primer sequence tails. The use of fluorescent labels allows for the direct visualization and quantification of the PCR products.
- Agarose Gel Electrophoresis: Agarose gel electrophoresis is the most commonly employed method for validating PCR results. This technique utilizes an electric current to separate DNA molecules based on their size. Agarose gels are used for DNA fragments larger than 500 base pairs, while polyacrylamide gels are used for smaller fragments. After electrophoresis, the DNA bands are visualized by staining the gel with DNA-specific dyes like ethidium bromide. The presence of a DNA band of the expected size, confirmed using a DNA ladder as a size reference, indicates successful amplification of the target sequence. The absence of any DNA bands suggests the absence of the target DNA, while the presence of incorrect size DNA bands indicates the production of unintended products.
- Restriction Enzyme Digestion: If direct DNA sequencing is not accessible or cost-effective, restriction enzyme digestion can be used as an indirect method to assess the sequence of the PCR amplicon. By selecting appropriate restriction enzymes that recognize specific DNA sequences within the amplicon, the presence or absence of expected restriction fragments can provide information about the integrity and specificity of the amplified product.
Validating PCR results using these techniques helps ensure the reliability and accuracy of the amplification process. These methods enable researchers to confirm the presence of the target sequence, identify any unintended products, and assess the overall success of the PCR experiment.
Types of PCR
PCR (Polymerase Chain Reaction) is a versatile technique that has undergone various modifications and adaptations to cater to specific needs in different fields. Here are some of the common types of PCR techniques:
- Conventional PCR: This is the standard PCR method where a single primer pair is used to bind to the two separated target strands. It generates millions of copies of the target DNA sequences.
- Multiplex PCR: This specialized PCR technique utilizes several pairs of primers that anneal to different target sequences in a single sample. It is commonly employed for the detection of pathogenic microorganisms and can identify mutations, deletions, insertions, and rearrangements in pathogenic specimens.
- Nested PCR: Nested PCR is used to enhance the specificity of DNA amplification by reducing nonspecific amplification. It involves two sets of primer pairs used in successive PCR reactions. The first round of PCR generates a larger DNA product that includes the target sequence, and the second PCR amplifies only the specific target sequence, improving specificity.
- Real-time PCR/Quantitative PCR (qPCR): This technique allows for the quantification of DNA amplification in real time during the PCR reaction. It is commonly used to estimate the number of DNA targets present in a sample or to study and compare gene expression. Real-time PCR can utilize nonspecific fluorescent dyes or sequence-specific DNA oligonucleotide fluorescent probes for amplification measurement.
- Hot Start/Cold Finish PCR: This PCR technique reduces nonspecific amplification during the initial stages of PCR. It involves using hybrid polymerases that remain inactive at ambient temperature and are only activated at higher temperatures. Inhibition of the polymerase activity at lower temperatures can be achieved using an antibody or covalently bound inhibitors.
- Touchdown PCR (Step-down PCR): Touchdown PCR is designed to minimize nonspecific amplification by gradually decreasing the primer annealing temperature in successive cycles. It starts with initial cycles having a higher annealing temperature for increased specificity and gradually reduces the temperature for more efficient amplification.
- Assembly PCR or Polymerase Cycling Assembly (PCA): This technique is used for the synthesis of long DNA molecules from long oligonucleotides with short overlapping segments. It involves an initial PCR with primers that have an overlap, followed by a second PCR using the products of the first PCR as templates to generate the final full-length DNA structure.
- Colony PCR: Colony PCR is a high-throughput technique used to confirm the presence of DNA inserts in recombinant clones. It amplifies the inserted sequences using specific primers designed for the vector regions flanking the insertion site. This technique allows for screening bacterial colonies transformed with recombinant vectors without initially extracting genomic DNA.
- Methylation-Specific PCR (MSP): MSP is a variant of PCR used to identify promoter hyper-methylation at CpG islands. It involves treating the target DNA with sodium bisulfite to convert unmethylated cytosine bases into uracil. Two types of primers, specific to methylated and unmethylated cytosine, are used to amplify the modified DNA, providing quantitative information about methylation when used in quantitative PCR.
- Inverse PCR: Inverse PCR is used to detect the sequences surrounding a target DNA (flanking sequences). It involves a series of restriction enzyme digestions and self-ligation. Primers are then used to amplify sequences at either end of the target DNA segment, extending outward from the known DNA segment.
- Reverse Transcription-PCR (RTP): RTP combines reverse transcription of RNA into complementary DNA (cDNA) using viral reverse transcriptase, followed by a conventional PCR using the resulting cDNA as a template. This technique is widely used in the detection of RNA viruses and the study of gene expression. A variant called differential-display reverse transcription-PCR or RNA arbitrarily primed PCR (RAP-PCR) allows for the comparison of gene expression under different conditions.
These different types of PCR techniques have expanded the applications of PCR in various fields, ranging from diagnostics and research to agriculture and environmental studies.
Variants of PCR
Variants of PCR (Polymerase Chain Reaction) have been developed to cater to various research, diagnostic, and industrial requirements. These variants offer specific advantages and are designed to address specific challenges encountered in traditional PCR techniques. Let’s explore some of the important variants of PCR:
- Extreme PCR: Extreme PCR involves increasing the concentration of primers and polymerase by 10-20 times. This amplification technique allows for rapid detection of virulent infectious and bioterrorism pathogens by achieving an amplification rate of 0.4-2.0 seconds per cycle.
- Photonic PCR: Photonic PCR utilizes fast heating and energy conversion to shorten the PCR time. This technique employs electronic resonance light-emitting diodes to achieve rapid energy conversion, resulting in target DNA amplification within 5 minutes. Photonic PCR offers greater convenience and speed in PCR detection.
- COLD-PCR: COLD-PCR stands for Co-amplification at Lower Denaturation temperature PCR. It is used to enrich mutant genes by reducing the reactive temperature of PCR. COLD-PCR takes advantage of the fact that DNA strands with base mismatches have lower denaturation temperatures compared to wild-type DNA. This variant is often employed in the detection of viral gene mutations, cancer-associated gene mutations, and other genetic variations.
- Nanoparticle-PCR: Nanoparticle-PCR involves the addition of gold nanoparticles to PCR reactions. Gold nanoparticles possess unique properties such as electrical, optical, thermal, and catalytic activities. By acting as additives in slowdown or touchdown PCR reaction systems, gold nanoparticles can significantly improve the amplification efficiency of high GC content templates. This variant is particularly useful for amplifying templates with high GC content.
- HPE-PCR: HPE-PCR, which stands for High G+C Content PCR, is designed for amplifying templates with long DNA chains and a high number of CTG repeats. It involves increasing the denaturation temperature of PCR to address the challenges posed by high G+C content templates .
- LATE-PCR: LATE-PCR (Linear-After-The-Exponential PCR) generates high concentrations of single-stranded DNA that can be analyzed at the endpoint using probes. This variant employs probes that hybridize over a wide temperature range, allowing for efficient and accurate analysis of amplified DNA.
- Digital PCR: Digital PCR (dPCR) is a technique that enables precise and sensitive quantification of nucleic acids. It utilizes two discrete optical channels for detection and focuses on the quantification of one or two targets within a single reaction. Digital PCR has gained popularity as a quantification strategy that combines absolute quantification with high sensitivity. It finds applications in healthcare and environmental analysis.
These variants of PCR demonstrate the versatility and adaptability of the PCR technique, allowing researchers and scientists to overcome specific challenges and achieve more accurate and efficient results in various fields of study.
Advantages of PCR
PCR offers several advantages that have made it a widely used and powerful technique in various fields. Here are some key advantages of PCR:
- Simplicity and speed: PCR is a relatively simple technique to understand and perform. It involves a few basic steps and can be carried out in a relatively short period of time, typically a few hours. This rapid turnaround time allows for quick diagnosis and identification of target sequences.
- High sensitivity: PCR is highly sensitive and has the potential to amplify even trace amounts of target DNA. It can produce millions to billions of copies of a specific DNA fragment, making it suitable for applications that require a high level of sensitivity, such as detecting rare genetic variants or low-abundance pathogens.
- Quantification capabilities: Real-time quantitative PCR (qPCR) is a variant of PCR that allows for the quantification of the synthesized product in real-time. This feature is particularly useful for analyzing alterations in gene expression levels, such as in tumors or microbial infections. qPCR enables researchers to accurately measure and compare gene expression levels across different samples.
- Versatility and flexibility: PCR is a versatile technique that can be used in various applications. It allows for the amplification of DNA from a wide range of sources, including genomic DNA, cDNA, and even degraded or old DNA samples. It can be adapted for different purposes, such as sequencing, cloning, mutation analysis, and genotyping.
- Specificity: PCR offers high specificity when designed properly. By using specific primers that target the desired DNA sequence, PCR can selectively amplify the target region while minimizing amplification of non-target DNA. This specificity ensures reliable and accurate results.
- Research tool: PCR is a powerful tool in research, enabling the sequencing of unknown disease etiologies and identification of new viral strains. It allows researchers to identify the sequence of previously unknown viruses related to known ones, providing a better understanding of diseases. PCR has greatly contributed to the advancement of genomics, molecular biology, and diagnostic research.
- Enhanced diagnosis and identification: PCR has significantly improved the diagnosis and identification of various diseases. It enables rapid and sensitive detection of pathogens, genetic disorders, and cancer-related mutations. PCR-based tests have become an integral part of clinical laboratories, providing faster and more accurate results for disease diagnosis.
Overall, PCR’s simplicity, speed, sensitivity, quantification capabilities, versatility, specificity, and its impact on research and diagnostics make it an indispensable tool in molecular biology, genetics, and clinical applications. Its continued development and refinement are expected to further enhance its applications and contribute to advancements in various scientific disciplines.
Limitations of PCR
While PCR has revolutionized biological sciences and enabled significant advancements, it also has certain limitations that need to be considered. Here are some limitations of PCR:
- Target sequence requirement: PCR requires prior knowledge of the target DNA sequence to design the primers for selective amplification. This means researchers need to know the specific sequence upstream of the target region on each single-stranded template. Without this information, PCR amplification cannot be performed.
- Potential for errors and mutations: Like all enzymes, DNA polymerases used in PCR are prone to errors during DNA synthesis, leading to mutations in the amplified fragments. These errors can introduce inaccuracies in the results and affect downstream analysis or interpretation.
- Contamination risks: PCR is highly sensitive, which is both an advantage and a limitation. Even a small amount of contaminating DNA, such as from previous PCR reactions or environmental sources, can be amplified and lead to false or ambiguous results. To minimize contamination risks, strict laboratory protocols should be followed, including separate rooms for reagent preparation, PCR setup, and analysis. Single-use aliquots and disposable pipettors should be used, and unidirectional workflow should be maintained.
- Inhibition by certain substances: PCR can be inhibited by the presence of certain chemicals such as ethanol, phenol, isopropanol, sodium dodecyl sulfate (SDS), high salt concentration, and chelators. These substances may be present in the sample or introduced during the experimental process, leading to failed or suboptimal amplification.
- Size limitations: There is an upper limit to the size of DNA that can be effectively amplified by PCR. Large DNA fragments may not be efficiently amplified or may require modified PCR protocols.
- Longer analysis time: While the PCR reaction itself is relatively quick, the analysis and detection of the PCR products often take longer. This can include gel electrophoresis, DNA sequencing, or other techniques to verify and analyze the amplified DNA. The additional time required for these steps should be considered in experimental planning.
- Inhibition by environmental samples: Environmental samples that contain humic acids, such as soil or water samples, can inhibit PCR amplification and result in inaccurate or failed results. Specialized purification methods or alternative PCR protocols may be required to overcome this limitation.
Awareness of these limitations is essential for researchers and practitioners working with PCR. By understanding these challenges, appropriate precautions can be taken to ensure the reliability and accuracy of PCR results in various applications.
Applications of Polymerase Chain Reaction (PCR)
The polymerase chain reaction (PCR) has revolutionized various fields of science and medicine due to its versatility and sensitivity. PCR and its advanced variants have made several applications possible, which were once considered impossible. Here are some key applications of PCR:
- Diagnosis of infections: PCR approaches are widely used in clinical laboratories to diagnose infections caused by bacteria, viruses, protozoa, and fungi. These techniques offer specific and sensitive detection and quantification of infectious agents.
- Diagnosis of genetic defects: PCR-based detection systems are used to accurately identify genetic disorders before the onset of disease and confirm their presence after the onset. PCR allows for the detection of inherited genetic changes and spontaneous genetic mutations.
- Diagnosis and prognosis of cancers: PCR-based approaches can identify cancer-related genes and analyze their expression patterns. This helps in determining genetic predisposition to certain types of cancer, confirming the cancer type, predicting prognosis, and guiding treatment decisions.
- Phylogenetics: PCR amplification of phylogenetic markers is routinely used in phylogenetic analysis to identify and classify organisms. It helps in understanding evolutionary relationships and biodiversity.
- Archeology: PCR techniques are employed to amplify and improve the quality and quantity of ancient DNA (aDNA) recovered from archaeological remains. This enables the analysis of aDNA for studying ancient populations, evolutionary history, and genetic diversity.
- Recombinant DNA technology: PCR is used in recombinant DNA technology to generate hybrid DNA molecules with precision. It is also employed to clone DNA into specific vectors for protein expression and production.
- Metagenomics: PCR is combined with metagenomics to identify rare genes and members of microbial communities. Gene-targeted metagenomics allows for the detection of rare species and rare genes present in complex microbial ecosystems.
- Site-directed mutagenesis: PCR-based approaches are commonly used to introduce mutations at specific locations in a gene. This technique helps in studying the role of specific amino acids in the structure and function of proteins.
- Personalized medicine: PCR technologies play a crucial role in pharmacogenomics and pharmacogenetics. Genetic markers tracked by PCR help determine individual responses to treatments, design tailored drugs, and prescribe effective drug doses.
- Forensic sciences: PCR is utilized in forensic sciences to amplify DNA samples obtained from crime scenes. Even poor-quality and low-quantity DNA samples can be reliably analyzed using PCR, aiding in criminal identification and investigations.
- DNA profiling: PCR-based methods are employed for DNA profiling, which exploits the polymorphic nature of DNA. These techniques help study ecological communities, phylogeny, population genetics, and provide valuable information in forensic investigations.
- Gene expression profiling: Reverse-transcriptase PCR and quantitative PCR (qPCR) are routinely used to analyze gene expression. They help in profiling gene expression patterns and validating transcriptome profiles obtained through techniques like microarray and RNA-seq.
- Identifying medicinal plants: PCR-based DNA barcoding is a rapid and accurate tool for identifying medicinal plant species. This approach is used in various fields, including medicine, ecology, and conservation biology, to identify endangered and new species.
- Detecting genetically modified organisms (GMOs): PCR techniques are employed to track the presence of genetically modified organisms in food and feed. This ensures their regulation and protects consumer rights by providing reliable and quick detection methods.
- Meat traceability: PCR methods are used to identify and quantify adulteration of meat in raw and processed food products. This helps ensure the accuracy and integrity of meat labeling and traceability systems.
The applications of PCR are extensive and continue to expand as new variants and techniques are developed. The technique’s sensitivity, specificity, and versatility have made it an indispensable tool in various scientific disciplines, clinical research, and diagnostic medicine.
Factors affecting Amplification
Sample Volume
- Generally, the amplification is performed at 20, 50, or 100 µl volume in 0.2 or 0.5 ml microfuge tubes.
- Larger volumes do not allow adequate thermal equilibrium of the reaction mixture.
Template DNA
- During PCR, generally, a nanogram amount of plasmid DNA, or microgram amount of genomic DNA is used.
- The higher amounts of template DNA can lead to the inhibition or result in non-specific amplification.
Primers
- Primers are synthetic oligonucleotides which are ranging from 15 to 30 bases.
- For PCR required a forward and a reverse primer. The melting temperature (Tm) of both primers is the same.
- The 3’ ends of Primers do not have more than two bases complementary to each other, as this helps in the formation of PRIMER-DIMER.
- The G+C content of Primers ranges from 40-60%.
- Low concentrations of primers result in poor yield of the specific product and high concentrations of primers may result in non-specific amplification. The optimal concentration of primers is between 0.1-1 µM.
Deoxynucleotide-tri-phosphates
- The final concentration of each dNTP (dATP, dGTP, dCTP & dTTP) in a standard amplification reaction is 200µM.
- It is important to keep the dNTP concentrations above the estimated Km of each dNTP (10 to 15 µM) for best base incorporation.
Taq DNA polymerase buffer
- 10X assay buffer is consist of 100 mM Tris- Cl (pH 9.0), 500 mM Potassium chloride, 15 mM MgCl 2 and 0.1% w/v gelatin.
- Mg2+ is an important cofactor needed for the activity of Taq DNA Polymerase. A low concentration of Magnesium will result in no amplification and high amounts may lead to the production of unwanted products.
Taq DNA Polymerase
- It is a 94 kD (Kilodaltons) thermostable DNA polymerase. The optimal temperature for Taq DNA Polymerase is 72°C.
- 3’ to 5’ exonuclease activity is absent but has 5’ to 3’ exonuclease activity and 5’ to 3’ polymerase activity.
- For most amplification reactions 1.5 to 2 units of the enzyme is recommended as higher enzyme concentration leads to non-specific amplification.
FAQ
What is PCR?
PCR is a laboratory technique used to amplify a specific DNA sequence, creating millions or billions of copies of the target DNA. It is based on the principles of DNA replication and involves a cyclic process of denaturation, annealing, and extension using DNA polymerase.
What is the purpose of PCR?
The primary purpose of PCR is to amplify a specific DNA sequence of interest. It is widely used in research, diagnostics, forensic analysis, genetic testing, and various other applications where the detection and analysis of DNA are necessary.
How does PCR work?
PCR involves repeated cycles of temperature changes to facilitate DNA denaturation, primer annealing, and DNA synthesis. The DNA sample is subjected to cycles of heating to separate the double-stranded DNA, cooling to allow primers to bind to the target sequence, and DNA synthesis by a heat-stable DNA polymerase.
What are the key components needed for PCR?
The essential components of a PCR reaction include a DNA template containing the target sequence, DNA primers that flank the target sequence, DNA polymerase (such as Taq polymerase), nucleotides (dNTPs), buffer solution, and magnesium ions (Mg2+).
What are the different types of PCR?
Some common types of PCR include conventional PCR, real-time PCR (qPCR), multiplex PCR, nested PCR, reverse transcription PCR (RT-PCR), and quantitative PCR (qPCR). Each type has specific variations and applications.
What is the role of primers in PCR?
Primers are short DNA sequences that bind to complementary regions flanking the target DNA sequence. They serve as starting points for DNA synthesis by the DNA polymerase. Primers are critical for target specificity and amplification.
What is the significance of PCR in genetic testing?
PCR is widely used in genetic testing to identify genetic mutations, detect infectious agents like viruses and bacteria, diagnose genetic diseases, determine gene expression levels, and perform DNA profiling for forensic analysis and paternity testing.
How can PCR be used in research?
PCR plays a crucial role in various research applications, including cloning, DNA sequencing, gene expression analysis, DNA fingerprinting, mutation detection, genotyping, and studying microbial diversity and evolution.
What are the limitations of PCR?
PCR has some limitations, such as the possibility of amplifying nonspecific DNA sequences if the primers are not specific enough, the potential for contamination leading to false results, the requirement for prior knowledge of the target sequence, and difficulties in amplifying highly GC-rich or repetitive DNA regions.
What is the role of PCR in the COVID-19 pandemic?
PCR has been instrumental in the diagnosis of COVID-19 by detecting the presence of SARS-CoV-2 viral RNA in patient samples. It has been widely used for mass testing, tracking the spread of the virus, and monitoring the effectiveness of vaccination campaigns.
References
- Liu, H. Y., Hopping, G. C., Vaidyanathan, U., Ronquillo, Y. C., Hoopes, P. C., & Moshirfar, M. (2019). Polymerase Chain Reaction and Its Application in the Diagnosis of Infectious Keratitis. Medical hypothesis, discovery & innovation ophthalmology journal, 8(3), 152–155.
- https://www.thermofisher.com/in/en/home/life-science/cloning/cloning-learning-center/invitrogen-school-of-molecular-biology/pcr-education/pcr-reagents-enzymes/pcr-applications.html
- https://himedialabs.com/TD/HTBM016.pdf
- http://www.ispybio.com/search/protocols/StudentPCR.pdf
- https://en.wikipedia.org/wiki/Polymerase_chain_reaction
- https://www.khanacademy.org/science/ap-biology/gene-expression-and-regulation/biotechnology/a/polymerase-chain-reaction-pcr
- https://microbenotes.com/polymerase-chain-reaction-pcr-principle-steps-applications/

