Table of Contents
What is Pulsed-field Gel Electrophoresis (PFGE)?
- Pulsed-field gel electrophoresis (PFGE) is a powerful technique used to separate large DNA molecules based on their size. It involves applying an electric field that periodically changes direction to a gel matrix, allowing for the effective separation of DNA molecules. This method has revolutionized various fields of research, including microbiology, epidemiology, and genetics.
- PFGE is particularly useful in microbiology for typing bacteria and studying their genetic similarity. By analyzing the DNA profiles obtained through PFGE, scientists can determine the relatedness of different bacterial strains. Similar species will exhibit similar profiles, while dissimilar ones will have distinct profiles. This feature is crucial in identifying the causative agent of a disease outbreak or tracing the source of contamination.
- The technique has also found applications in monitoring and evaluating microorganisms in clinical samples, soil, and water. By analyzing the DNA patterns of microorganisms present in these samples, researchers can gain valuable insights into the prevalence, spread, and diversity of microbial populations. PFGE has become a reliable and standard method in vaccine preparation, helping to ensure the quality and consistency of vaccines.
- The discovery of PFGE can be attributed to the efforts of several pioneering scientists in the late 1970s and early 1980s. In 1977, Dr. David Burke and colleagues at the University of Colorado developed a method for separating DNA molecules based on size using conventional gel electrophoresis. They laid the foundation for the subsequent development of PFGE. In 1983, Dr. Richard L. Sweeley and colleagues at the DuPont Company coined the term “pulsed-field gel electrophoresis” and described a technique to separate large DNA molecules by applying alternating electric fields to a gel matrix.
- Over the years, researchers such as Dr. J. Peter Gerhardt at the University of Alabama in Birmingham and Dr. Mark Scherer at the University of Minnesota further refined and improved the technique. PFGE became widely adopted in the late 1980s and early 1990s for the separation and analysis of large DNA molecules in various areas of molecular biology, genetics, and biotechnology.
- PFGE’s versatility and reliability have made it an indispensable tool in controlling, preventing, and monitoring diseases in different populations. Its ability to accurately determine genetic relatedness has proven crucial in outbreak investigations, epidemiological studies, and the development of effective prevention strategies. With ongoing advancements in molecular biology techniques, PFGE continues to evolve and contribute to our understanding of DNA, genetics, and microbial populations.
Principle of Pulsed Field Gel Electrophoresis (PFGE)
The principle of pulsed-field gel electrophoresis (PFGE) revolves around the separation of DNA fragments based on their size using an electric field. In conventional gel electrophoresis, small DNA fragments migrate more easily through the gel matrix than large fragments. However, when dealing with DNA fragments larger than 30 to 50 kilobases (kb), a threshold length is reached where all large fragments run at the same rate and appear as a single diffuse band on the gel.
To overcome this limitation, PFGE introduces a crucial element—the periodic changing of the electric field direction. By periodically reversing the field direction, the different lengths of DNA fragments react at different rates to the change. Larger DNA fragments are slower to realign their charge when the field direction is switched, while smaller fragments respond more quickly. This dynamic change in field direction causes the DNA fragments to separate progressively over time, even at very large lengths.
As a result, PFGE enables the separation of extremely large DNA fragments that would otherwise appear as a single band in conventional gel electrophoresis. The consistent alternating of the electric field’s direction during PFGE allows for the distinct separation and visualization of individual DNA bands corresponding to various fragment lengths. This principle forms the basis for the effective separation and analysis of large DNA molecules in PFGE, making it a valuable technique in various areas of research, including microbiology, genetics, and biotechnology.
Types of Pulsed Field Gel Electrophoresis
Pulsed Field Gel Electrophoresis (PFGE) is a powerful technique used for the isolation and typing of large DNA molecules. Different types of PFGE methods have been developed based on the specific requirements of the study and the available resources. Here are some of the commonly used types of PFGE:
- Contour Clamped Homogeneous Electric Field (CHEF): CHEF is a widely used PFGE system that allows precise control over the size, location, coordination, stability, and continuity of the electric field. The system uses a complex method to create distortion on the edge of the chamber and passive electrodes. All the electrodes in the CHEF system are connected to the power supply through a series of identical ring resistors, which regulate the voltage on the electrodes. The voltage is set at 24 points, and the system uses a hexagonal arrangement of 24 electrodes. CHEF is capable of separating DNA molecules over 7000 kb in size.
- Field-Inversion Gel Electrophoresis (FIGE): FIGE was introduced in 1986 and is known for its ability to separate smaller DNA fragments. It involves the use of two fields with separated angles that periodically turn upside down. Shorter time pulses or less traction are used for the forward movement of molecules, while the reverse movement is achieved by longer time pulses. It should be noted that the movement of DNA molecules larger than 2 Mb may not always be consistent, and molecules of different sizes and labels can sometimes migrate together in FIGE. Nevertheless, FIGE provides acceptable resolution and can separate DNA fragments over 800 kb.
- Asymmetric Field Inversion Gel Electrophoresis (AFIGE): AFIGE is specifically used for the detection of DNA double-strand breaks (DSBs). It involves exposing DNA to an inverted and asymmetric electric field for a longer period of time, which increases the occurrence of DNA breakage. This method is particularly suitable for the detection of double-stranded DNA.
- Orthogonal-Field Alternation Gel Electrophoresis (OFAGE): OFAGE is a vertical alternating field electrophoresis method that utilizes regular spot electrodes. It creates nonlinear and dissimilar electric fields to change the final migration pattern of DNA samples. The major drawback of this method is the use of non-unit electric fields, which results in variable migration patterns depending on the gel situation. However, OFAGE is capable of separating DNA molecules between 1000 kb and 2000 kb in size.
- Rotating Gel Electrophoresis (RGE): RGE was introduced by Southern in 1987 and is characterized by the rotation of the gel between two angles while the power supply is turned off. This method utilizes a monotonous electric field and direct separation occurs with the use of only one set of electrodes. RGE is suitable for the separation of DNA fragments ranging from 50 kb to 6000 kb.
- Pulsed-Homogeneous Orthogonal Field Gel Electrophoresis (PHOGE): PHOGE differs from other PFGE methods with homogeneous fields in its orientation angle of 90 degrees in the electric field. DNA molecules are moved four times per cycle instead of twice, resulting in DNA lines that do not follow a straight path due to the use of multiple electric fields. PHOGE is capable of separating DNA fragments over 1 Mb in size.
- Programmable Autonomously-Controlled Electrodes (PACE): PACE is an advanced electrophoresis system that allows independent adjustment of voltage on 24 electrodes arranged in a closed contour. This system provides flexibility in producing controlled homogeneous electric fields, voltage gradients, and flow duration. PACE is capable of separating DNA fragments ranging from 100 bp to over 6 Mb. The ability to change the angle between the reorientation of alternating currents enables faster separation of large DNA molecules compared to other alternating electrophoresis methods.
These various types of PFGE methods offer researchers different options for isolating and typing large DNA molecules based on their specific needs and resources available.
How does Pulsed-field Gel Electrophoresis work?
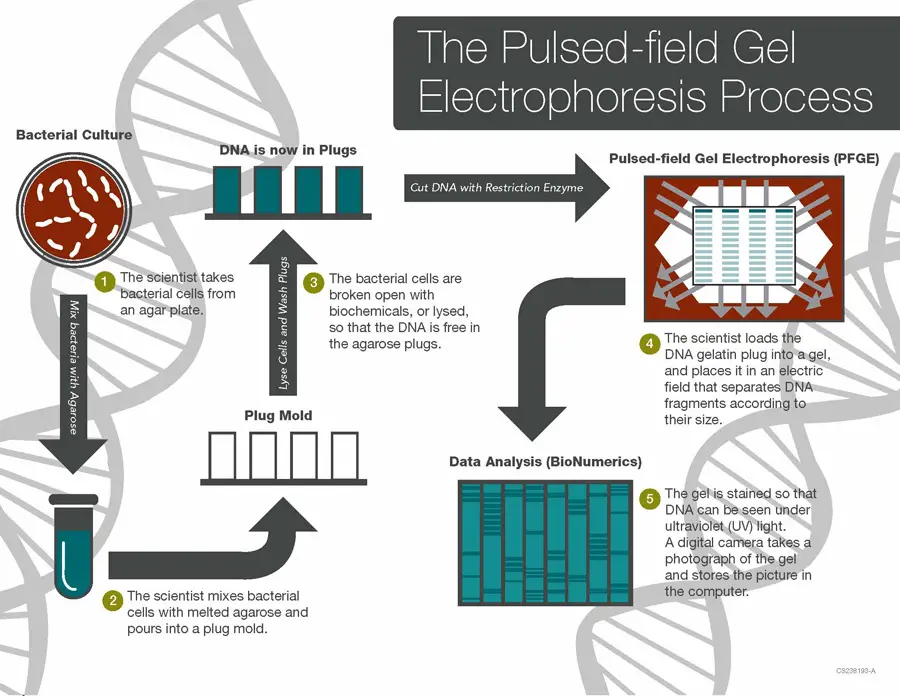
Pulsed-field Gel Electrophoresis (PFGE) is a technique used to separate and analyze large fragments of DNA. Here is a step-by-step explanation of how PFGE works:
- Extraction: The process begins by extracting bacterial cells from an agar plate. These cells are typically mixed with agarose, a gel-like substance, which will serve as the medium for DNA separation.
- Gel preparation: The bacterial cells and agarose mixture is poured into a mold to form plugs. These plugs contain the bacterial cells embedded in the solidified agarose matrix.
- Cell lysis: The bacterial cells within the agarose plugs are lysed, or broken up, using biochemicals or other methods. This step releases the DNA from the plugs, allowing it to be accessible for analysis.
- Gel loading: The DNA gel plugs are loaded into the gel electrophoresis apparatus, usually positioned in an alternating checkerboard pattern. This arrangement ensures that the electric field is applied in different directions during the separation process.
- Electrophoresis: An electric field is applied to the gel, causing the DNA fragments to migrate through the gel matrix. The unique feature of PFGE is that the electric field is pulsed, meaning it alternates in direction and intensity at regular intervals. This pulsed field enables the separation of very large DNA fragments.
- Staining and visualization: After electrophoresis, the gel is stained with a DNA-specific dye that allows the DNA fragments to be visualized under ultraviolet (UV) light. The fragments appear as distinct bands on the gel, creating a DNA fingerprint.
- Image analysis: The gel is photographed using a digital camera, and the image is saved on a computer. The DNA fingerprint, represented by the banding pattern, is analyzed using specialized software such as BioNumerics. This software aids in comparing and analyzing the fingerprint patterns.
- Database comparison: The DNA fingerprint generated from PFGE is compared to a database of known fingerprints, such as the national database managed by PulseNet Central. Database managers examine the patterns to determine if they match known outbreaks or are part of an ongoing epidemic.
- Further investigation: If a match or potential outbreak is identified, microbiologists and epidemiologists from public health agencies collaborate to investigate the outbreak further. This may involve conducting additional tests, tracking cases, and implementing control measures to contain the spread of the disease.
Method of Pulsed Field Gel Electrophoresis (PFGE)
- The process is similar to the procedure used for the standard electrophoresis, except that instead of running the voltage in a single direction, the voltage is frequently changed between three directions: one running through the central direction of the gel’s axis and two running in a 60-degree angle to either from the other.
- The pulse times are the same for each direction , which results to a net movement of DNA.
The major steps involved in Pulsed-field gel electrophoresis are
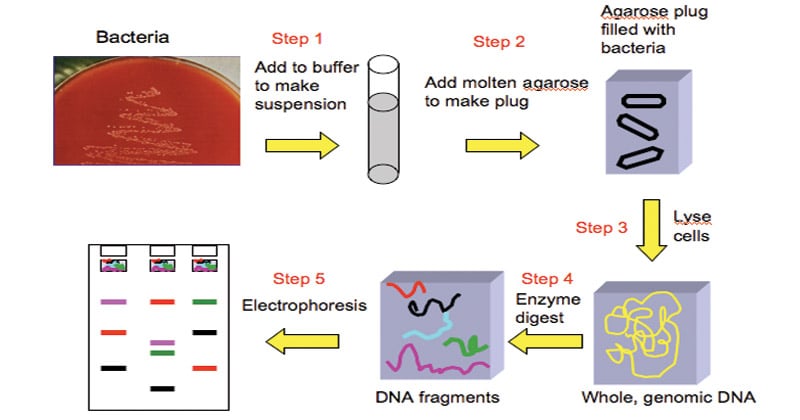
The major steps involved in Pulsed-field gel electrophoresis (PFGE) are as follows:
- Preparing the agarose gel: Agarose powder is mixed with TBE buffer and heated to create an agarose gel. This gel is poured into a casting tray and allowed to solidify. The gel will serve as the medium through which the DNA fragments will migrate during electrophoresis.
- Loading the DNA sample: The DNA sample is digested with a specific restriction enzyme, which cuts the DNA at specific recognition sites. This digestion generates a set of DNA fragments of known sizes. The digested DNA sample is loaded into wells created in the gel. Each well contains a specific DNA sample that will be separated based on fragment size.
- Running the electrophoresis: The gel is placed in an electrophoresis chamber filled with TBE buffer, which provides the appropriate conditions for the electrophoresis process. An electric field is applied to the gel in a pulsed manner. The direction of the electric field is changed at regular intervals to allow the DNA fragments to reorient themselves. This pulsing of the electric field helps in separating large DNA fragments.
- Staining the gel: After the electrophoresis is complete, the gel is removed from the chamber. The gel is then stained with a DNA-specific stain, such as ethidium bromide, which binds to the DNA and allows for its visualization. Ethidium bromide-stained DNA fluoresces when exposed to UV light. The stained gel is observed under UV light, and the DNA fragments appear as distinct bands. These bands represent different-sized DNA fragments, with larger fragments migrating more slowly through the gel.
By following these steps, PFGE allows for the separation and visualization of DNA fragments based on their size, enabling the analysis of genome structure, genetic relatedness, and other applications in molecular biology and epidemiology.
Components to be enhanced while performing PFGE for the best results
To enhance the performance of Pulsed Field Gel Electrophoresis (PFGE) and achieve optimal results, attention should be given to the following components:
- Voltage: The voltage applied during PFGE plays a crucial role in the separation of DNA fragments. For molecules of a few hundred kilobases (kb) in size, a standard voltage of 6 V/cm is typically used. However, when dealing with larger sample sizes in megabases (Mb), it is recommended to use a lower voltage. Conversely, for smaller sample sizes, a higher voltage can be employed to enhance resolution.
- Pulse angle: The pulse angle refers to the angle at which the electric field switches its direction during the electrophoresis process. The commonly used angle is 120°, but it can be adjusted based on the size of the DNA fragments being analyzed. To improve the resolution of larger fragments, smaller angles can be used. Conversely, larger angles are suitable for smaller fragments.
- Switch time: Switch time refers to the duration for which the electric current is applied in one direction before switching direction. It is important to select an appropriate switch time depending on the size of the DNA fragments. Short switch times are suitable for smaller fragment sizes, while longer switch times are beneficial for larger fragment sizes.
- Temperature: PFGE runs are typically conducted over an extended period, often overnight or several days. It is crucial to maintain the buffer temperature consistently throughout the procedure. Lower temperatures result in longer run times but provide greater resolution, while higher temperatures reduce run time but may yield lower resolution. Careful control of the temperature ensures optimal results.
By optimizing these key components, researchers can fine-tune the PFGE parameters to achieve the best possible separation and resolution of DNA fragments. It is essential to consider the specific characteristics of the samples and adjust these parameters accordingly to enhance the overall performance of PFGE.
Applications of Pulsed Field Gel Electrophoresis (PFGE)
Pulsed Field Gel Electrophoresis (PFGE) has a wide range of applications in various areas of genetic and genomic research. Some of the major applications of PFGE include:
- Construction of Physical Maps: PFGE enables the separation and characterization of large DNA fragments, allowing the construction of physical maps for the chromosomes of bacterial species. This provides valuable information about the size and arrangement of genetic material in the genome.
- Genotyping and Genetic Fingerprinting: PFGE is commonly used for genotyping and genetic fingerprinting of organisms. By analyzing the banding patterns generated through PFGE, researchers can identify genetic variations and distinguish between different strains or isolates of a species.
- Epidemiological Studies: PFGE is considered a gold standard in epidemiological studies of pathogenic organisms. It helps in tracking and investigating disease outbreaks by comparing the DNA fingerprints of isolates obtained from different sources such as clinical samples, food, or the environment. This information aids in understanding the transmission and spread of infectious agents.
- Subtyping of Pathogens: PFGE subtyping has proven particularly useful in discriminating among strains of pathogens such as Listeria monocytogenes. It enables the identification of specific strains associated with clinical infections and helps establish links between environmental or food isolates and disease cases.
- Genome Structural Mapping: PFGE is utilized to plot structural maps of bacterial genomes. Large DNA fragments are digested with restriction enzymes, allowing segregation and separation of different parts of the genome. This information helps in understanding the size, shape (circular or linear), and organization of bacterial genomes.
- Genetic Mapping: PFGE-generated structural maps can be used for genetic mapping. By hybridizing cloned genes from the separated fragments, researchers can create genetic maps that provide insights into the arrangement and organization of genes within the genome.
- Identification of Antibiotic Resistance: PFGE can assist in the identification of antibiotic-resistant strains, such as methicillin-resistant Staphylococcus aureus (MRSA) strains. By comparing the DNA fingerprints of different isolates, researchers can determine the presence of specific resistance markers.
- Classification and Taxonomy: PFGE is employed in the classification and taxonomy of bacteria. It helps in differentiating between closely related species or strains by analyzing their unique DNA fingerprint patterns.
- Other Applications: PFGE has additional applications, including aetiologic studies of bacteria, identification of infectious agents in clinical or environmental samples, analysis of large DNA fragments, detection of DNA viruses with large genomes, determination of genome size and chromosomal mechanisms in eukaryotes and prokaryotes, preparation of Yeast Artificial Chromosome (YAC) libraries, and transgenic mouse studies.
Advantages of Pulsed Field Gel Electrophoresis (PFGE)
Pulsed Field Gel Electrophoresis (PFGE) offers several advantages that make it a valuable tool in genetic and genomic research:
- Wide Size Range: PFGE is capable of separating DNA fragments ranging from a few kilobase pairs (kb) to over 10 megabase pairs (Mb). This broad size range allows for the analysis and characterization of large DNA fragments, including whole chromosomes and large genomic regions.
- High Discrimination Power: PFGE subtyping has proven to be highly effective in differentiating between strains of various pathogenic bacteria. It exhibits a high concordance with epidemiological relatedness, meaning that DNA fingerprints generated through PFGE accurately reflect the genetic variations and relatedness among bacterial isolates.
- Enhanced Discrimination: PFGE has been shown to be more discriminating than other typing methods such as ribotyping or multi-locus sequence typing (MLST) for many bacteria. It can provide a higher level of resolution and better differentiate between closely related strains.
- Universal Application: PFGE, in its basic format, can serve as a universal and generic method for subtyping bacteria. By choosing the appropriate restriction enzyme and optimizing the electrophoresis conditions, PFGE can be applied to a wide range of bacterial species without requiring significant modifications.
- Stability and Reproducibility: DNA restriction patterns generated by PFGE are known for their stability and reproducibility. This means that the same bacterial isolate analyzed at different times or by different laboratories is likely to yield consistent results. This reliability is crucial in epidemiological studies and outbreak investigations.
- Compatibility with DNA Libraries: PFGE is compatible with the preparation of DNA libraries, such as Yeast Artificial Chromosome (YAC) libraries. The ability to separate and analyze large DNA fragments makes PFGE a valuable tool for constructing genomic libraries and studying specific genetic elements.
- Long-Range Structural Information: PFGE provides valuable insights into the organization and structure of genomes. By separating DNA fragments based on their size and arrangement, PFGE aids in the construction of physical maps and the characterization of large genomic regions.
- Versatile and Established Technique: PFGE is a well-established technique with a long history of successful applications in genetic and genomic research. It is widely used in various fields, including epidemiology, microbiology, genetics, and genomics, and has been refined and optimized over time.
In summary, the advantages of PFGE include its ability to separate DNA fragments across a wide size range, its high discriminatory power, its universality as a subtyping method, the stability and reproducibility of DNA patterns, its compatibility with DNA libraries, the provision of long-range structural information, and its status as a versatile and well-established technique. These advantages make PFGE an invaluable tool for various applications in genetic and genomic research.
Limitations of Pulsed Field Gel Electrophoresis (PFGE)
While Pulsed Field Gel Electrophoresis (PFGE) offers valuable insights, it also has some limitations that should be considered:
- Time-consuming: PFGE is a labor-intensive technique that requires several steps, including DNA extraction, lysis, digestion, and electrophoresis. The process can be time-consuming, particularly when analyzing a large number of samples.
- Requirement for trained technicians: PFGE requires trained and skilled technicians who are familiar with the technique and its nuances. Proper execution and interpretation of results depend on the expertise of the personnel involved.
- Limited discrimination power: Although PFGE is highly discriminatory for many bacteria, it may not differentiate between all unrelated isolates. Some strains may have similar or identical restriction patterns, leading to challenges in distinguishing between them using PFGE alone.
- Variability between technicians: Even with standardized protocols, slight variations in technique or interpretation can occur between different technicians. This can introduce some level of inconsistency or variation in the generated DNA patterns.
- Inability to optimize separation uniformly: PFGE does not allow for simultaneous optimization of separation throughout the entire gel. Variations in gel conditions, such as voltage and running time, may result in different resolution or separation efficiencies for different regions of the gel.
- Ambiguity regarding DNA band identity: Although bands of the same size are often assumed to represent the same DNA fragment, PFGE does not provide definitive information about the identity of these bands. Further analyses, such as DNA sequencing or hybridization, may be required to confirm the composition of specific bands.
- Lack of independence among bands: Bands observed in PFGE patterns are not independent entities. Changes in a single restriction site can result in alterations in multiple bands, making it challenging to assign specific genetic events or rearrangements based solely on band patterns.
- Limited phylogenetic information: While PFGE patterns can indicate relatedness among strains, they do not provide a comprehensive phylogenetic measure. PFGE should be considered as a tool for guiding relatedness assessments rather than a definitive method for establishing evolutionary relationships.
- Inability to type certain strains: Some bacterial strains may not be effectively typed using PFGE due to technical limitations or specific characteristics of the organism. These strains may require alternative typing methods to achieve meaningful results.
Considering these limitations, it is important to carefully interpret PFGE results, complement them with additional analyses when necessary, and be aware of the potential constraints and challenges associated with the technique.
FAQ
What is Pulsed-field Gel Electrophoresis (PFGE)?
PFGE is a technique used to separate and analyze large DNA fragments, typically ranging from a few kilobases to several megabases, based on their size using an electric field.
What is the purpose of PFGE?
PFGE is primarily used for molecular fingerprinting and genotyping of bacteria, allowing researchers to study genetic relatedness, identify outbreaks, and trace the source of infections.
How does PFGE differ from traditional gel electrophoresis?
Unlike traditional gel electrophoresis, PFGE uses alternating electric fields at regular intervals to separate large DNA fragments, providing enhanced resolution and the ability to analyze fragments that are too large for conventional methods.
What are the key steps involved in PFGE?
The major steps in PFGE include preparing the agarose gel, loading the DNA sample, running the electrophoresis with pulsed electric fields, and staining the gel to visualize the separated DNA fragments.
What are the advantages of PFGE?
PFGE allows for the separation of large DNA fragments, provides high resolution, is versatile for different bacterial species, and offers stable and reproducible results.
What are the limitations of PFGE?
Some limitations of PFGE include being time-consuming, requiring trained technicians, limited discrimination between unrelated isolates, slight result variations between technicians, and the inability to type certain strains.
Can PFGE be used for genotyping other organisms besides bacteria?
Yes, PFGE can be used for genotyping other organisms with large genomes, such as fungi, protozoa, and even mammals. It is a versatile technique for analyzing the structure and origin of genomes.
Is PFGE a widely accepted technique in epidemiological studies?
Yes, PFGE is considered a gold standard in epidemiological studies of pathogenic organisms due to its high discriminatory power and ability to link isolates from outbreaks and infections.
What factors can be optimized to enhance PFGE results?
Factors that can be optimized for better PFGE results include voltage, pulse angle, switch time, and temperature, depending on the size of the DNA fragments being analyzed.
Can PFGE be combined with other molecular techniques?
Yes, PFGE can be combined with other techniques such as DNA sequencing, PCR, and hybridization methods to further analyze and characterize DNA fragments of interest.
References
- Sharma-Kuinkel BK, Rude TH, Fowler VG Jr. Pulse Field Gel Electrophoresis. Methods Mol Biol. 2016;1373:117-30. doi: 10.1007/7651_2014_191. PMID: 25682374; PMCID: PMC4582012.
- Parizad EG, Parizad EG, Valizadeh A. The Application of Pulsed Field Gel Electrophoresis in Clinical Studies. J Clin Diagn Res. 2016 Jan;10(1):DE01-4. doi: 10.7860/JCDR/2016/15718.7043. Epub 2016 Jan 1. PMID: 26894068; PMCID: PMC4740595.
- https://thesciencenotes.com/pulsed-field-gel-electrophoresis-pfge/
- https://www.bio-rad.com/en-in/applications-technologies/pulsed-field-gel-electrophoresis?ID=LUSORPDFX
- https://www.sciencedirect.com/topics/medicine-and-dentistry/pulsed-field-gel-electrophoresis
- https://www.applied-maths.com/applications/pulsed-field-gel-electrophoresis-pfge-typing
- https://microbeonline.com/pulsed-field-gel-electrophoresis-pfge/
- https://international.neb.com/tools-and-resources/video-library/what-is-pulsed-field-gel-electrophoresis
- https://experiments.springernature.com/articles/10.1385/0-89603-498-4:33
