Table of Contents
What is Real-Time PCR?
- Real-time polymerase chain reaction (PCR), also known as quantitative PCR (qPCR), is a variant of the PCR technique that allows for the detection and quantification of amplified DNA or RNA in real-time during the amplification process. It was developed as an advancement to traditional PCR methods and has become an invaluable tool in various fields of research, including molecular biology and genetics.
- The principle behind real-time PCR is similar to that of traditional PCR, where specific segments of DNA or RNA are amplified enzymatically in vitro. However, real-time PCR incorporates the use of specialized fluorescent dyes or probes that enable the continuous monitoring and measurement of the amplification process as it occurs.
- During real-time PCR, the DNA or RNA sample of interest is mixed with primers, nucleotides, and a DNA polymerase enzyme, similar to traditional PCR. Additionally, fluorescent dyes or probes are included, which bind specifically to the amplified DNA or RNA sequences. These dyes or probes emit fluorescence signals when they are excited by a specific wavelength of light.
- The real-time PCR instrument consists of a thermal cycler that can cycle through multiple temperature stages rapidly. It can heat and cool the reaction mixture at precise intervals to facilitate the denaturation, annealing, and extension steps of the amplification process. As the reaction progresses, the DNA or RNA is denatured into single strands, the primers bind to their complementary sequences, and the DNA polymerase enzyme extends the primers to synthesize new DNA strands.
- The fluorescence emitted by the dyes or probes is detected by the real-time PCR instrument after each amplification cycle. The instrument measures the increase in fluorescence intensity, which is directly proportional to the amount of amplified DNA or RNA present in the reaction. By comparing the fluorescence signals to a standard curve generated using known concentrations of a target molecule, the initial amount of the target in the sample can be determined.
- Real-time PCR offers several advantages over traditional PCR methods. Firstly, it provides quantitative data, allowing researchers to measure the exact amount of the target DNA or RNA in the sample. This is particularly useful for gene expression studies, pathogen detection, viral load determination, and genetic variation analysis.
- Furthermore, real-time PCR eliminates the need for post-amplification analysis, such as gel electrophoresis, as the fluorescence signals are captured and analyzed in real-time. This saves time and reduces the risk of contamination or handling errors associated with additional steps.
- Overall, real-time PCR has revolutionized the field of nucleic acid amplification and analysis. Its ability to provide accurate, sensitive, and quantitative results in a rapid and automated manner has made it an indispensable tool in various scientific disciplines.
Definition of Real-Time PCR
Real-time PCR, also known as quantitative PCR (qPCR), is a molecular biology technique that allows for the amplification, detection, and quantification of DNA or RNA in real-time during the amplification process. It uses fluorescent dyes or probes to measure the increase in fluorescence intensity, providing quantitative data on the amount of target nucleic acid present in the sample.
Objectives of RT-PCR
- The objectives of real-time PCR (RT-PCR) are twofold: amplifying specific segments of RNA and utilizing this amplification for various applications such as diagnosing infections, studying genes, and analyzing gene expression.
- The primary objective of RT-PCR is to amplify a specific segment of RNA. By using reverse transcriptase enzyme, the RNA is converted into complementary DNA (cDNA) during the initial reverse transcription step. This cDNA then serves as a template for amplification in the PCR process. Through a series of heating and cooling cycles in a thermal cycler, the DNA polymerase enzyme amplifies the target RNA segment, resulting in the production of billions of copies of the desired RNA segment.
- One important application of RT-PCR is the diagnosis of certain infections. By targeting specific RNA sequences associated with pathogens or viral genomes, RT-PCR can detect the presence of these infectious agents in clinical samples. The high sensitivity and specificity of RT-PCR make it a valuable tool for identifying and diagnosing infections, including viral diseases like COVID-19.
- In addition to infection diagnosis, RT-PCR is extensively used for studying genes and gene expression. Researchers can analyze the mRNA or microRNA levels present in a sample to gain insights into gene expression patterns. By quantifying the amount of RNA of interest, RT-PCR enables researchers to investigate the expression of specific genes under various conditions, such as during disease progression or in response to drug treatments. This information contributes to our understanding of gene regulation, cellular processes, and disease mechanisms.
- In summary, the objectives of RT-PCR are to amplify specific RNA segments and utilize this amplification for diagnosing infections, studying genes, and analyzing gene expression. By achieving these objectives, RT-PCR plays a crucial role in various areas of research and diagnostics, facilitating advances in molecular biology and genetics.
Real Time PCR Principle
The Principle of Real-Time PCR, also known as quantitative PCR (qPCR), is a powerful technique used to amplify and quantify specific DNA or RNA sequences in real-time. It builds upon the principles of reverse transcription PCR (RT-PCR) and traditional PCR.
In real-time PCR, the process begins by converting the RNA sample into complementary DNA (cDNA) using an enzyme called reverse transcriptase. This step is known as reverse transcription. The reverse transcriptase enzyme synthesizes a complementary DNA strand using the RNA template as a guide. As a result, the RNA is converted into double-stranded cDNA.
Once the cDNA is obtained, it can be further amplified and quantified using the principles of PCR. The cDNA is heated to denature or separate the double-stranded DNA into two single-stranded DNA templates. These templates contain regions that are complementary to specific primers.
Primers are short DNA sequences that are designed to bind to specific target regions within the DNA sequence of interest. These primers are designed based on the principles of nucleic acid hybridization, where complementary sequences will anneal or bind to each other.
During the annealing step of real-time PCR, the temperature is lowered, allowing the primers to specifically bind to their complementary sequences on the single-stranded DNA templates. This step ensures that the primers are targeted to the desired DNA sequence, which will be amplified.
After annealing, the temperature is increased, and DNA polymerase, an enzyme responsible for DNA replication, extends the primers by adding nucleotides to the 3′ end of each primer. This step is known as elongation, and it results in the synthesis of a complementary DNA strand using the original DNA template.
The three steps of denaturation, annealing, and elongation are repeated multiple times in a cyclic manner within a specialized thermal cycler machine. This machine controls and regulates the temperature changes required for each step. By cycling through these temperature changes, millions of copies of the cDNA are produced.
During the amplification process, the real-time PCR machine also detects and quantifies the amount of amplified DNA in real-time. This is achieved by incorporating a fluorescent dye or probe into the PCR reaction. As DNA is synthesized, the dye binds to the newly synthesized DNA strands and emits fluorescence. The fluorescence signal is measured at each cycle, allowing the quantification of the DNA or RNA of interest.
Real-time PCR offers several advantages over traditional PCR methods. It enables the detection and quantification of specific DNA or RNA sequences in real-time, without the need for post-amplification analysis. This makes it a valuable tool in various applications, including gene expression analysis, pathogen detection, and genetic testing.
In summary, the principle of real-time PCR combines reverse transcription and PCR techniques to amplify and quantify specific DNA or RNA sequences. By leveraging the principles of nucleic acid hybridization and DNA replication, real-time PCR allows for the amplification and measurement of target sequences in real-time, providing valuable insights into various biological processes.
Requirements (Enzymes) of RT-PCR
RT-PCR, or reverse transcription PCR, requires several enzymes and other components to facilitate the conversion of RNA into cDNA and subsequent amplification. The key requirements for RT-PCR include:
- Nucleic Acid Sample (Sample RNA): The starting material for RT-PCR is RNA, specifically the RNA of interest that contains the target sequence to be amplified. This RNA can be isolated from various sources, and in most cases, mRNA is used as the sample.
- Reverse Transcriptase Enzyme: Reverse transcriptase is a vital enzyme in RT-PCR. It catalyzes the synthesis of complementary DNA (cDNA) strands from the RNA template. This enzyme is also known as RNA-dependent DNA polymerase because it uses the RNA strand as a template to produce the complementary DNA strand. Reverse transcriptase is responsible for the central dogma reversal in which genetic information flows from RNA to DNA. It is the key enzyme that converts the sample RNA into cDNA, which can then be amplified.
- DNA Polymerase Enzyme: Once the cDNA is generated from the RNA template, a DNA polymerase enzyme is used for the subsequent amplification steps in PCR. The most commonly used DNA polymerase in PCR, including RT-PCR, is Taq DNA polymerase. Taq DNA polymerase is derived from the bacterium Thermus aquaticus and is known for its thermostability. It can withstand the repeated cycles of heating and cooling during PCR and continue its activity.
- Primers: Primers are short DNA sequences that are complementary to specific regions of the target sequence. In RT-PCR, three types of primers are commonly used:
- Random Primers: These short single-stranded sequences of 6 to 8 nucleotides can bind to the complementary sites of RNAs, with or without a poly(A) tail. They are used for cDNA synthesis using reverse transcriptase.
- Oligo(dT) Primers: Oligo(dT) primers are oligonucleotides typically consisting of 12 to 18 nucleotides, with a segment of repeating deoxythymidine (dT). They bind specifically to the poly(A) tail present at the 3′ end of mRNA molecules.
- Sequence-specific Primers: These short single-stranded sequences of nucleotides are designed to bind to specific regions of the target RNA sequence. They are used in one-step RT-PCR, where reverse transcription and PCR amplification occur in a single reaction.
- Deoxynucleotide Triphosphates (dNTPs): dNTPs are the building blocks for DNA synthesis. They are artificially synthesized nucleotides that are incorporated by DNA polymerase during DNA strand elongation. The four different dNTPs used in PCR are dATP, dGTP, dTTP, and dCTP, representing the four nucleotide bases.
- PCR Buffers and Other Chemicals: PCR buffers containing salts, pH stabilizers, and other components are used to maintain the optimal conditions for enzyme activity during the RT-PCR process. Additional chemicals, such as magnesium ions, may also be included to support the enzymatic reactions.
- Thermocycler (PCR Machine): A thermocycler, commonly known as a PCR machine, is required to perform the temperature cycling necessary for the denaturation, annealing, and extension steps of PCR. It allows precise control of temperature changes and cycling times, ensuring the proper execution of each step in the RT-PCR process.
In summary, RT-PCR requires enzymes such as reverse transcriptase and DNA polymerase, along with specific primers, dNTPs, buffers, and a thermocycler. These components work together to convert RNA into cDNA and amplify the target sequence, enabling the detection and quantification of specific RNA molecules in a sample.
Real-Time PCR Protocol
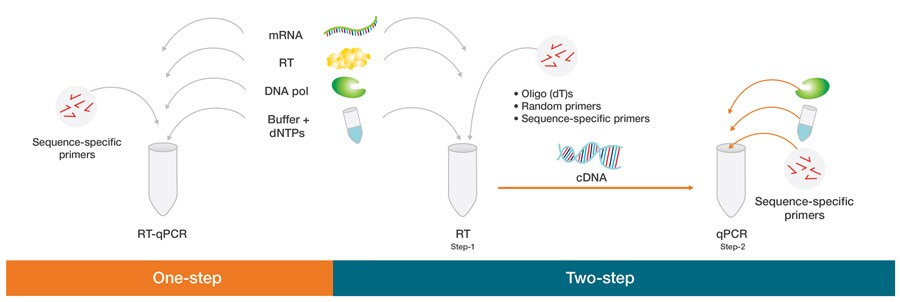
It is a valuable tool, use for evaluating gene expression by estimating the abundance of certain RNAs. There are two methods are available for Real-Time PCR such as; one-step RT-qPCR and two-step RT-qPCR.
In both method, there is a common step where the RNA is first reverse-transcribed into cDNA, after that this cDNA is used as the template for qPCR amplification.
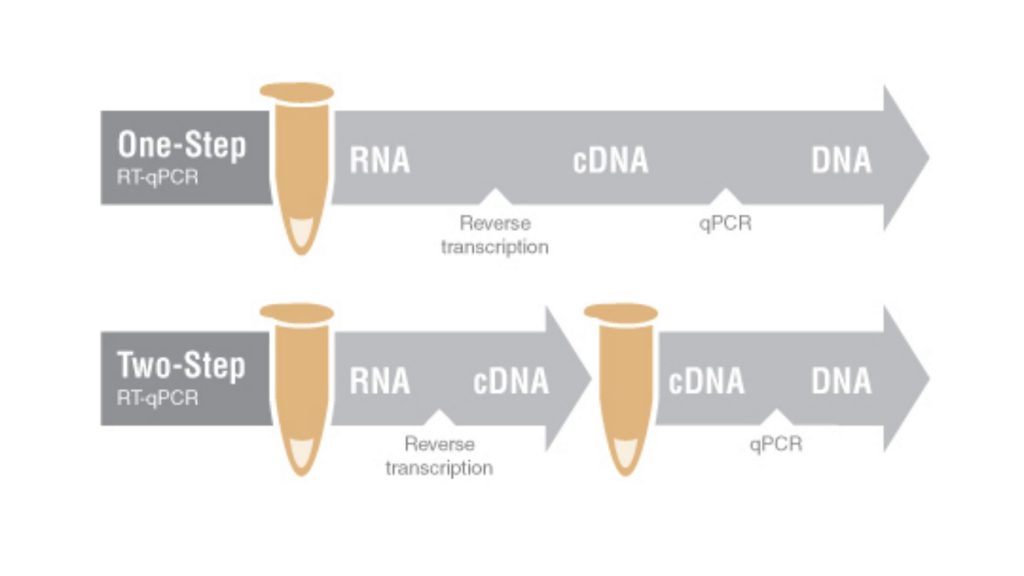
In one-step RT-qPCR, cDNA synthesis and qPCR are conducted within a single reaction vessel in a common reaction buffer. While in two-step RT-qPCR, cDNA is synthesized in one reaction, and an aliquot of the cDNA is then used for a subsequent qPCR experiment.
One-Step RT-qPCR
One-Step RT-qPCR is a molecular biology technique that combines the reverse transcription (RT) and quantitative polymerase chain reaction (qPCR) processes in a single reaction. It offers several advantages and is commonly used in gene expression analysis, viral detection, and other applications where RNA quantification is required. Here are the key points about One-Step RT-qPCR:
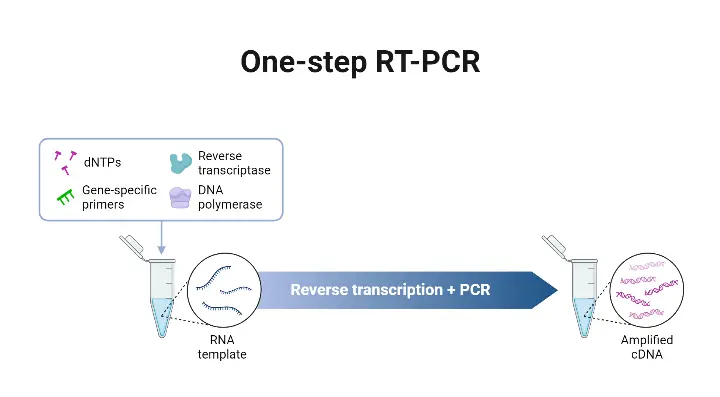
- Single Reaction Vessel and Common Buffer: In this method, a single reaction vessel and a common buffer are used to carry out both cDNA synthesis and qPCR amplification. This simplifies the experimental setup and reduces the risk of contamination.
- Suitable for High Sample Size or Limited Known Targets: One-Step RT-qPCR is particularly useful when dealing with a large number of samples or a limited number of known targets that need to be repeatedly amplified. It streamlines the workflow and reduces the overall hands-on time.
- Gene-Specific Primers: One-Step RT-qPCR utilizes gene-specific primers that anneal to specific target sequences in the RNA template. These primers are designed to anneal at higher temperatures compared to random primers. The use of gene-specific primers improves the specificity and accuracy of the assay.
- Higher RT Reaction Temperatures: In some One-Step RT-qPCR protocols, higher RT reaction temperatures are used compared to traditional two-step workflows. This can enhance the efficiency of reverse transcription and cDNA synthesis. Engineered or novel reverse transcriptases (RTs) are sometimes employed in One-Step RT-qPCR, as they can withstand higher reaction temperatures and improve the overall performance of the assay.
One-Step RT-qPCR offers a streamlined and efficient approach for gene expression analysis and RNA quantification. By combining reverse transcription and qPCR in a single reaction, it simplifies the experimental workflow and reduces the risk of experimental variability. It is particularly advantageous when working with a large number of samples or when amplifying a limited number of known targets repeatedly. The use of gene-specific primers and the potential for higher RT reaction temperatures contribute to improved specificity, accuracy, and efficiency in One-Step RT-qPCR assays.

Advantage of One-Step RT-qPCR
One-Step RT-qPCR, also known as one-step reverse transcription quantitative PCR, offers several advantages over other PCR methods. Some of the key advantages of one-step RT-qPCR are:
- Simple and Easy Handling Setup: One-step RT-qPCR has a simplified setup compared to multi-step RT-PCR methods. It combines reverse transcription and quantitative PCR into a single reaction, reducing the number of steps involved. This simplicity makes it easier to set up and perform the experiment, even for those with limited experience in molecular biology techniques.
- Higher Accuracy and Specificity: One-step RT-qPCR provides higher accuracy and specificity in target sequence amplification. The use of specific primers and fluorescent probes allows for precise detection and quantification of the target RNA or DNA sequence. The inclusion of reverse transcription in the same reaction ensures that the starting template is accurately converted into cDNA before amplification.
- Lesser Chance of Contamination: Since one-step RT-qPCR involves fewer pipetting steps, there is a reduced chance of errors and contamination. Each pipetting step introduces the possibility of introducing contaminants or inaccuracies. By minimizing these steps, one-step RT-qPCR helps maintain the integrity of the samples and reduces the risk of cross-contamination between reactions.
- Cheaper and Faster Method: One-step RT-qPCR is a cost-effective and time-efficient method. It eliminates the need for a separate reverse transcription reaction, which reduces the cost of reagents and labor. Additionally, by combining the reverse transcription and amplification steps into a single reaction, the overall processing time is reduced, allowing for faster results.
- Improved Data Reproducibility: One-step RT-qPCR offers improved data reproducibility due to its closed-tube reactions. The risk of sample evaporation or contamination during sample transfer is minimized since all the necessary components are present in a single tube. This ensures consistency between replicates and improves the reliability of the results.
- Suitable for High-Throughput Applications: One-step RT-qPCR is well-suited for high-throughput applications where multiple samples need to be processed simultaneously. Its streamlined workflow and reduced sample handling make it efficient for large-scale experiments. It allows researchers to analyze a significant number of samples in a shorter amount of time.
- Minimal Sample Handling and Reduced Bench Time: With one-step RT-qPCR, the sample handling is minimized, reducing the chances of sample loss or degradation. The reverse transcription and amplification steps are performed in a single tube, eliminating the need for intermediate transfers. This not only saves time but also reduces the risk of sample contamination or mix-up. The reduced bench time increases the efficiency of the laboratory workflow.
- Fast and Highly Reproducible: One-step RT-qPCR provides fast and highly reproducible results. The combined reverse transcription and amplification steps enable a streamlined process, reducing the overall reaction time. The reliability and consistency of the results are enhanced due to the minimized variations introduced by separate reaction steps.
In summary, one-step RT-qPCR offers advantages such as simple setup, higher accuracy and specificity, reduced chances of contamination, cost and time savings, improved data reproducibility, suitability for high-throughput applications, minimal sample handling, and fast and highly reproducible results. These benefits make one-step RT-qPCR a valuable tool in various molecular biology and diagnostic applications.
Disadvantages of One-Step RT-qPCR
One-Step RT-qPCR, despite its advantages, has certain disadvantages that should be taken into consideration. These disadvantages include:
- Requirement for New RNA Samples: One-step RT-qPCR requires new RNA samples to examine new targets or repeat experiments. Unlike two-step RT-qPCR, where cDNA can be stored and used for further analysis, one-step RT-qPCR does not allow for the storage of cDNA formed during the reaction. Therefore, if additional targets need to be examined or if repeat experiments are required, new RNA samples need to be obtained.
- Detection of Fewer Targets per Sample: Due to the combination of reverse transcription and amplification into a single reaction, one-step RT-qPCR has a limitation in the number of targets that can be detected per sample. The reaction conditions are a compromise between the two reactions, making it difficult to optimize the reverse transcription and amplification separately. As a result, the detection of multiple targets becomes more challenging.
- Less Sensitivity Compared to Two-Step RT-qPCR: One-step RT-qPCR is generally less sensitive compared to the two-step approach. The combined reaction conditions in one-step RT-qPCR, which include reverse transcription and amplification, can result in a compromise in reaction conditions. This compromise may lead to reduced sensitivity and the detection of fewer templates per reaction mixture.
- Higher Chance of Primer-Dimer and Non-Specific Binding: Due to the combination of multiple chemicals and components in a single reaction tube, one-step RT-qPCR has a higher chance of primer-dimer formation and non-specific binding. This can result in background noise and false-positive signals, affecting the specificity and accuracy of the assay.
- Higher Chance of Reaction Failure: Compared to two-step RT-qPCR, one-step RT-qPCR has a relatively higher chance of reaction failure. The complex nature of the combined reaction, with multiple components and steps, increases the likelihood of experimental variability and technical difficulties. Factors such as suboptimal primer design, suboptimal reaction conditions, or issues with reverse transcriptase and DNA polymerase activity can contribute to reaction failure.
- Larger Template Requirement: Due to the lower template detection capacity in one-step RT-qPCR, a larger amount of starting template is often required to achieve reliable and detectable results. This can be a limitation when working with limited or precious samples.
It’s important to consider these disadvantages when deciding on the appropriate PCR method for a specific experiment. While one-step RT-qPCR offers advantages in terms of simplicity and speed, its limitations should be carefully evaluated to ensure the desired outcomes can be achieved.
Standard Protocol of One-Step RT-qPCR
The standard protocol for performing One-Step RT-qPCR involves the following steps:
- Prepare the Reaction Mix: In sterile RNase-free microfuge tubes, mix the following components, except for the RNA template:
- OneTaq One-Step Reaction Mix (2X): 25 μl
- OneTaq One-Step Enzyme Mix (25X): 2 μl
- Gene-specific Forward Primer (10 μM): 2 μl
- Gene-specific Reverse Primer (10 μM): 2 μl
- Nuclease-free H2O: 19–x μl (adjust to reach a final volume of 50 μl)
- Add RNA Template: Add the RNA template to the reaction mix last. The recommended amount of RNA is up to 1 μg, and the volume is denoted as x μl. This will vary depending on the specific experiment and the desired concentration of RNA template.
- Start Reactions Immediately: Once the RNA template is added, start the reactions immediately to minimize the risk of RNA degradation. The reaction mix should be thoroughly mixed to ensure proper dispersion of all components.
- Cycling Parameters: The One-Step RT-qPCR reaction typically consists of the following cycling steps:
- Reverse Transcription: Incubate at 48°C for 15–30 minutes. This step allows the reverse transcriptase enzyme to convert RNA into cDNA.
- Initial Denaturation: Perform an initial denaturation step at 94°C for 1 minute. This denatures the cDNA and prepares it for subsequent amplification.
- Denaturation, Annealing, and Extension: Repeat the following three steps for the desired number of cycles:
- Denaturation: Heat to 94°C for 15 seconds. This step separates the DNA strands.
- Annealing: Lower the temperature to the specific annealing temperature (usually between 50°C and 65°C) for 30 seconds. The primers anneal to their target sequences on the template DNA.
- Extension: Increase the temperature to 68°C for 1 minute per kilobase (kb) of the expected amplicon size. The DNA polymerase extends the primers and synthesizes new DNA strands.
- Final Extension: Perform a final extension step at 68°C for 5 minutes to ensure completion of any remaining DNA synthesis.
- Hold: After the cycling steps are completed, the reaction is typically held at 4°C indefinitely or until further analysis or storage.
| CYCLE STEP | TEMPERATURE | TIME | CYCLES |
| Reverse Transcription | 48°C | 15–30 minutes | 1 |
| Initial Denaturation | 94°C | 1 minute | 1 |
| Denaturation Annealing Extension | 94°C 50–65°C 68°C | 15 seconds 30 seconds 1 minute per kb | 40 |
| Final Extension | 68°C | 5 minutes | 1 |
| Hold | 4°C | ∞ | 1 |
Two-step RT-qPCR
Two-step RT-qPCR is a widely used molecular biology technique that involves two separate steps: reverse transcription (RT) and quantitative polymerase chain reaction (qPCR). This method offers flexibility and advantages in certain experimental scenarios. Here are the key points about Two-step RT-qPCR:
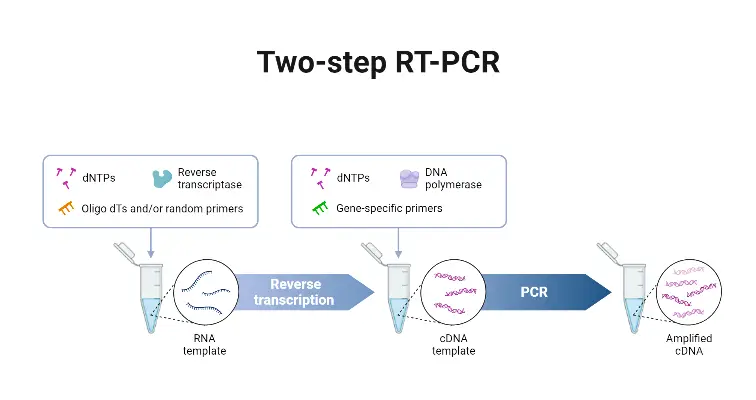
- cDNA Synthesis with Random Hexamers: In the first step of Two-step RT-qPCR, random hexamers are used as primers to carry out the synthesis of complementary DNA (cDNA). These random hexamers can be a mixture of oligo-dT primers, which anneal to the poly(A) tails of mRNA, and/or gene-specific primers, which target specific gene sequences. The use of random hexamers allows for the generation of cDNA from a mixture of various RNA species present in the sample.
- Suitable for Precious Samples and Multiple Targets: Two-step RT-qPCR is preferred when the specimen is precious or when there is a need to examine multiple targets. This method allows flexibility to test new targets by going back to the saved cDNA or using a portion of the cDNA produced during the RT step. It is also useful when the targets of interest are not well characterized and may require further testing.
- cDNA as a Mixture of RNA Species: The cDNA synthesized during the RT step of Two-step RT-qPCR represents a mixture of various RNA species present in the sample. This includes mRNA transcripts from different genes, providing a comprehensive representation of the transcriptome at the time of RNA extraction.
- Flexibility in cDNA Handling: In Two-step RT-qPCR, the cDNA synthesis reaction can be scaled up to accommodate higher RNA input. Additionally, extraction and precipitation steps can be employed to concentrate and further purify the cDNA if required. This flexibility allows researchers to obtain higher yields of cDNA and optimize its quality for downstream qPCR analysis.
- Multiple Applications of cDNA: The produced cDNA can be used not only in the immediate qPCR step but also saved for future use. This allows for the quantification of the expression of multiple genes from a single RNA/cDNA sample, providing an efficient use of the generated cDNA.
Two-step RT-qPCR offers versatility and convenience in gene expression analysis and transcript quantification. By separating the reverse transcription and qPCR steps, researchers have more flexibility in handling precious samples, examining multiple targets, and characterizing new targets. The cDNA generated during the RT step can be stored and used for subsequent qPCR experiments or quantification of gene expression from the same sample.
Advantages of Two-step RT-qPCR
Two-step RT-qPCR, a widely used molecular biology technique, offers several advantages in gene expression analysis and transcript quantification. Here are the key advantages of Two-step RT-qPCR:
- Storage of cDNA: One of the notable advantages of the two-step method is the ability to store the cDNA formed during reverse transcription. This allows for future use and provides the opportunity to quantify gene expression from the same RNA/cDNA sample at a later time. It enables researchers to analyze multiple targets or perform additional experiments without repeating the reverse transcription step.
- Higher Efficiency, Accuracy, and Reliability: Two-step RT-qPCR generally exhibits higher efficiency, accuracy, and reliability compared to other methods. It allows for the detection of larger templates per reaction mixture, increasing the sensitivity of the assay. This makes it suitable for experiments requiring precise and reliable quantification of gene expression levels.
- Reduced Chance of Reaction Failure and Non-specific Binding: Two-step RT-qPCR has a lower chance of reaction failure, non-specific binding, and primer-dimer formation. By separating the reverse transcription and PCR steps, potential issues related to primer-dimer formation and non-specific binding are minimized, leading to more accurate and specific results.
- Flexibility in Enzyme Selection: Two-step RT-qPCR provides greater flexibility in selecting reverse transcriptase (RT) enzymes and DNA polymerases for the PCR step separately. This allows researchers to optimize the choice of enzymes based on specific experimental requirements, such as the need for higher RT reaction temperatures or specialized DNA polymerases for challenging templates.
- Suitable for Limited Starting Material: The two-step method is the approved approach for applications involving a limited amount of starting material, such as single-cell analysis. It allows for efficient cDNA synthesis from small quantities of RNA, enabling gene expression analysis at the single-cell level.
Overall, Two-step RT-qPCR offers advantages in terms of flexibility, efficiency, accuracy, and reliability. The ability to store cDNA for future use, reduced chances of reaction failure and non-specific binding, and the flexibility in enzyme selection make it a preferred method for gene expression analysis and transcript quantification in various research applications.
Disadvantages of Two-step RT-qPCR
Two-step RT-qPCR, while offering advantages, also has some disadvantages that should be considered. Here are the key disadvantages of Two-step RT-qPCR:
- Extra Open-Tube Step: One of the notable disadvantages of the two-step method is the requirement for an extra open-tube step. This additional step introduces a potential source of contamination during the transfer of cDNA from the reverse transcription reaction to the qPCR reaction. It increases the risk of introducing external contaminants, which can affect the accuracy and reliability of the results.
- Increased Pipetting Manipulations: Two-step RT-qPCR involves more pipetting manipulations compared to other methods. The multiple pipetting steps required for the transfer of samples and reagents between tubes increase the likelihood of variability and potential contamination. Each pipetting step introduces the risk of error or cross-contamination, which can impact the reliability of the experimental results.
- Longer Hands-On Time: The two-step method generally requires a longer hands-on time compared to other RT-qPCR protocols. The separation of the reverse transcription and qPCR steps extends the overall experimental duration. This increased hands-on time can be a limiting factor when processing a large number of samples or when rapid results are desired.
- Higher Chance of Contamination: Due to the multiple pipetting manipulations and open-tube transfer steps involved, there is a higher chance of contamination in the two-step RT-qPCR method. Contamination can lead to inaccurate and unreliable results, affecting the interpretation of gene expression data. Stringent laboratory practices and precautions must be followed to minimize the risk of contamination.
- Complex and Resource-Intensive Process: Two-step RT-qPCR is a more complex and resource-intensive process compared to other RT-qPCR workflows. It requires more reagents, tubes, and equipment, increasing the overall cost of the experiment. Additionally, the method may require a well-trained person to perform the technique accurately and effectively. The complexity and resource requirements can limit its feasibility in certain laboratory settings or experimental contexts.
It is important to consider these disadvantages and assess the specific experimental requirements and constraints when deciding on the appropriate RT-qPCR protocol. While two-step RT-qPCR has its drawbacks, it continues to be a widely used method due to its suitability for certain applications and the ability to store cDNA for further analysis.
Protocol of Two-step RT-qPCR
Step One: Reverse Transcription
Reagents Required
- Reverse Transcriptase MMLV-RT (100 units/µl)
- 10X RT Buffer (500 mM Tris-HCl, pH 8.3, 750 mM KCl, 30mM MgCl2 50 mM DTT)
- Random decamers (50µM)
- dNTP (2.5mM each dNTP)
- RNase Inhibitor (10 units/µl)
Place RNase Inhibitor and Reverse Transcriptase ON ICE directly from the Box. Thaw 10x reaction buffer, random decamers, and dNTP mix quickly in your hands and place ON ICE;
Procedure
- Use small 0.25ml PCR tubes
- Assemble your reaction as follows on ice. Add the enzyme last.
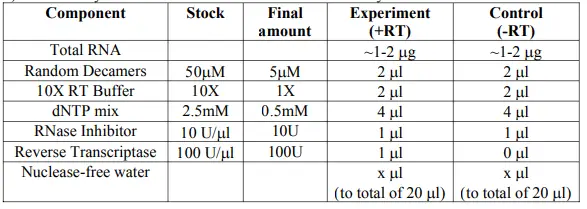
- Mix gently, spin briefly.
- Incubate in the thermocycler at: a. 44°C for 1 hr; b. 92°C for 10 min to inactivate the reverse transcriptase.
- Store reaction at –20°C or proceed to the PCR.
Step Two: PCR
A. Primer Preparation
- Primers are shipped in dry form. Briefly spin the tube before you open the cap to avoid loss of DNA pellet.
- Dissolve the oligonucleotide in 10mM Tris, pH7.5 to make a primer stock at 100µM concentration.
- Dilute from this stock 1:20 (in water) to make a working solution at 5 µM for use in setting up PCR reactions.
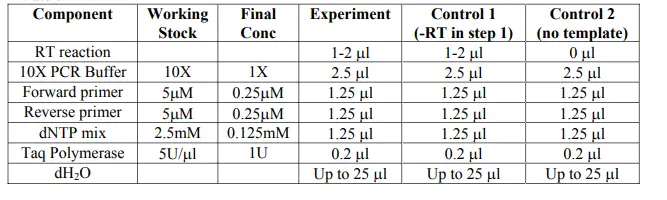
B. Setting up PCR reactions
Negative Controls
Use two negative controls among the PCRs.
- The minus-RT control from the previous step, or alternatively, untreated RNA can simply be subjected to PCR.
- A minus-template PCR, should have all the PCR components but use water as a template instead of an aliquot of the cDNA (RT reaction). This control will verify that none of the PCR reagents is contaminated with DNA.
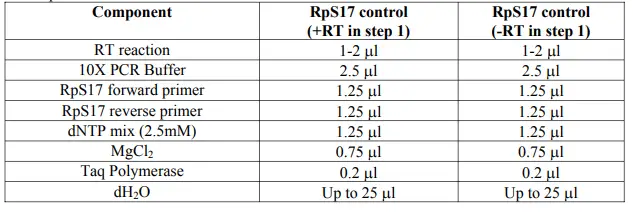
Positive Control
- Perform PCR to amplify a cDNA that corresponds to a basal/housekeeping transcript, e.g., ribosomal protein S17 (RpS17). The TA’s will provide each team with RpS17 primers. For your reference, these are the primer sequences: Forward RpS17: 5′ – cga acc aag acg gtg aag aag – 3′ Reverse RpS17: 5′ – cct gca act tga tgg aga tac c – 3′
Expected RT-PCR product size: 211bp .The primers are located on different exons that are separated by a 59bp intron. If genomic DNA is amplified, the product size would be 270bp.
- Use genomic DNA isolated from S2 cells as template. If your primers span intron(s), note the size of the expected PCR product and if necessary, adjust annealing temperature of the PCR program.
Reagents:
- Taq DNA polymerase: Invitrogen
- With 10X Buffer (-MgCl2) and 50mM MgCl2
The following table outlines the components needed for PCR. Note: do not use DEPC-treated water.
For RpS17 control:
Making Master Mixes:
- Consider making master mixes if you are testing multiple sets of primers at once. A master mix will contain everything except the PCR primers. If you are testing n sets of primers, make a master mix enough for n+1 tests.
- Mix the components gently but thoroughly. Aliquot 22.5µl of your master mix to each tube.
- Add 1.25µl of each of the appropriate primers at 5µM working stock concentration.
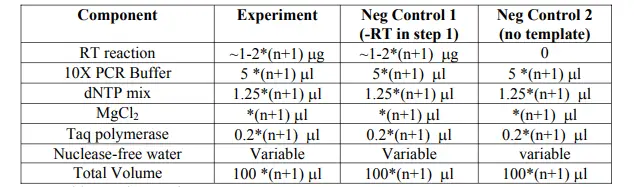
- Assemble reactions on ice.
Incubate in Thermacycler:
- Initial denaturation: 94°C for 4 min
- 30 cycles: Denature at 94°C for 30 sec, Anneal at 55°C for 20–30 sec**, Extend at 72°C for 45 sec ***
- Final extension: 72°C for 5 min
Note: Start with the annealing temperature suggested by your primer design software. An annealing temperature of ~55°C used with the cycling times shown is often a reasonable starting point, but the optimal temperature and cycling times for your primer and template combination may need to be determined empirically.
Note: The rule of thumb is to use an extension time of 1 min per kilobase of the target.
Run 15 µl of your reaction on 1-1.5% Agarose gel
General Steps/Procedure of RT-PCR
The general steps/procedure of RT-PCR can be summarized into four stages: the preparatory stage, reverse transcription, amplification, and product analysis stage. Here’s an overview of each stage:
- Preparatory Stage: In this stage, the necessary materials and reagents are prepared for the RT-PCR reaction. This involves arranging all the materials, ensuring safety measures are followed, cleaning the PCR reaction preparation area, and bringing the RNA sample from storage or extracting it. For one-step RT-PCR, all the reaction components, including sample RNA, reverse transcriptase enzyme, RNase H, primers, DNA polymerase, dNTPs, buffers, and other components, are added to a single reaction tube. In two-step RT-PCR, the sample RNA, reverse transcriptase, RNase H, primers, dNTPs, and other buffers and chemicals for reverse transcription are loaded into a tube.
- Reverse Transcription: In this primary step, the RNA is converted into complementary DNA (cDNA) using reverse transcriptase enzyme. In one-step RT-PCR, the reverse transcription occurs directly in the same reaction tube where all the components are added. The tube is subjected to a specified temperature (usually 40-50°C) for a specific time (10 to 30 minutes) in a thermocycler. During this step, the primers bind to the respective sites on the RNA sample, and the reverse transcriptase synthesizes cDNA by adding free dNTPs.
- Amplification: The amplification step is similar to the process in other PCR techniques for DNA amplification. In one-step RT-PCR, the same reaction mixture from the reverse transcription step undergoes the amplification process. In two-step RT-PCR, the cDNA is isolated and transferred to another tube where DNA polymerase, primers, PCR buffer, dNTPs, and other chemicals are added. The tube is then placed in a thermocycler for amplification. This step involves cycles of denaturation, annealing, and elongation, which are repeated for a specific number of cycles determined by the user.
- Product Analysis Stage: In this final stage, the reaction mixture subjected to PCR is analyzed to confirm the desired amplification. Gel electrophoresis is commonly used for product analysis, where the amplified products are separated based on size in an agarose gel and visualized under UV light. However, in real-time RT-PCR, there is no need for this additional step. The fluorescence emitted by the reaction mixture during amplification is continuously monitored using specialized equipment, allowing for real-time detection and quantification of the amplification products.
These general steps provide an overview of the RT-PCR procedure, which can be modified based on specific experimental requirements or variations in the one-step and two-step RT-PCR methods.
Quantitation of RT-PCR Results
Quantitating the results obtained from real-time reverse transcription polymerase chain reaction (RT-PCR) experiments is essential for understanding gene expression levels and comparing different samples. Two commonly used strategies for quantitation are the standard curve method and the comparative threshold (Ct) method. Let’s explore these methods:
1. Standard Curve Method
The standard curve method involves constructing a standard curve using an RNA or DNA of known concentration. This curve serves as a reference for quantifying mRNA targets of unknown concentrations. While RNA standards can be used, they may introduce variability due to their stability and the need for in vitro transcription. Alternatively, other nucleic acid samples such as purified plasmid DNA or cDNA samples expressing the target gene can be used. These samples can be quantified using spectrophotometric measurements and converted into copy number values.
It’s important to note that using cDNA plasmids as standards will only provide information on relative changes in mRNA expression, as they do not control for variations in the efficiency of the reverse transcription step. To account for this and variability in RNA inputs, normalization to a housekeeping gene is recommended.
2. Comparative Ct Method
The comparative Ct method, also known as the 2–ΔΔCt method, compares the Ct values of the samples of interest with a control or calibrator. The Ct values of both the calibrator and the samples are normalized to an endogenous housekeeping gene, which serves as an internal control.
To calculate ΔΔCt, the Ct value of the samples is subtracted from the Ct value of the calibrator for both the target gene and the reference gene. The formula is:
ΔΔCt = ΔCt_sample – ΔCt_reference
If the amplification efficiencies of the target gene and the reference gene are approximately equal, the ΔΔCt calculation is valid. This can be determined by examining how ΔCt varies with template dilution. If the plot of cDNA dilution versus ΔCt is close to zero, it indicates similar amplification efficiencies. However, if a suitable housekeeping gene with similar amplification efficiency cannot be found, the standard curve method is preferred.
In summary, the standard curve method utilizes a reference curve constructed from known concentrations, while the comparative Ct method compares Ct values between samples and a calibrator. Both methods have their advantages and considerations, and the choice depends on the experimental setup and available resources.
Fluorescence Markers for Real-Time PCR
Fluorescence markers play a crucial role in real-time PCR for the detection and quantification of amplified DNA or RNA. Two commonly used fluorescence markers in real-time PCR are Taqman Probe and SYBR Green. Here’s some information about these markers:
1. Taqman Probe
Taqman Probe is a hydrolysis probe used in real-time PCR. It consists of an oligonucleotide with a reporter dye, often fluorescein (FAM), attached to the 5′ end, and a quencher dye, such as tetramethylrhodamine (TAMRA), attached to the 3′ end. The probe is designed to be complementary to a specific target sequence within the DNA or RNA being amplified.
During the amplification process, the Taq polymerase enzyme binds to the target gene sequence and starts extending the DNA strand. As the extension occurs, the 5′ nuclease activity of the Taq polymerase degrades the Taqman Probe. This degradation separates the reporter dye from the quencher, allowing the reporter dye to emit fluorescence. The increase in fluorescence intensity is directly proportional to the amplification of the target gene, providing quantitative information about the target.
2. SYBR Green
SYBR Green is a fluorescent dye that binds to the minor groove of double-stranded DNA. When bound to DNA, SYBR Green emits a strong fluorescent signal. It has higher signal intensity compared to other dyes like Ethidium Bromide or Acridine Orange.
SYBR Green is advantageous because it provides data about each cycle of amplification and can also be used for analyzing the melting temperature of the amplified product. This makes it a popular choice for real-time PCR assays. However, one of the disadvantages of SYBR Green is its lack of specificity compared to Taqman Probe. SYBR Green binds to any double-stranded DNA, so it may produce non-specific signals from unintended PCR products or primer dimers.
In summary, Taqman Probe offers higher specificity due to its probe-target hybridization and degradation mechanism, while SYBR Green provides more flexibility in data analysis but with the risk of non-specific signal detection. The choice of fluorescence marker depends on the specific requirements of the experiment, including sensitivity, specificity, and data analysis needs.
Advantages of Real-Time PCR
Real-time PCR (RT-PCR) offers several advantages over traditional PCR methods. Here are the key advantages of real-time PCR:
- Rapid and Efficient Amplification: Real-time PCR enables the rapid and enzymatic amplification of RNA or DNA samples, producing millions of copies in a short period. It allows for fast and efficient analysis of nucleic acids, saving time in the laboratory workflow.
- Automation and Ease of Use: The process of real-time PCR is semi-automatic and regulated by a thermocycler, reducing the need for extensive manual intervention. This automation makes it a simple and user-friendly technique, ensuring reproducibility and minimizing human error.
- High Specificity and Sensitivity: Real-time PCR offers high specificity and sensitivity, allowing for the precise detection and quantification of target RNA or DNA molecules. The technique can accurately identify RNA viruses and classify them at the strain level. It has revolutionized the identification and diagnosis of viral infections by shortening the time required for analysis.
- Detection of Minute Amounts of Target RNA: Real-time PCR can detect very small amounts of target RNA, often as low as 5 picograms. This sensitivity is significantly higher compared to traditional methods like Northern Blot. It enables the detection of rare transcripts or low-abundance RNA molecules, enhancing the sensitivity of gene expression studies.
- Study of Mutated Genes and Gene Expression: Real-time PCR allows for the easy and prompt study of mutated genes and gene expression patterns. It has facilitated the early-stage diagnosis of cancer, monitoring the results of gene therapy, and investigating gene insertion experiments. The technique provides accurate and quantitative data on gene expression levels.
- Qualitative and Quantitative Analysis: Real-time PCR is a versatile method that can be used for both qualitative and quantitative analysis of RNA or DNA samples. It enables the identification and quantification of target molecules in the same reaction, providing valuable information about the presence, abundance, and dynamics of specific nucleic acids.
- Wide Dynamic Range and High Analytical Sensitivity: Real-time PCR offers a wide dynamic range of quantification, typically spanning 7-8 logarithmic decades. It allows for the detection and quantification of a broad range of target concentrations. The technique also exhibits high analytical sensitivity, capable of detecting as few as 5 copies or 1 femtogram to 10 picograms of DNA per PCR reaction.
- Closed System and Reduced Contamination Risk: Real-time PCR is performed in a closed system, reducing the risk of contamination. The use of specialized reaction tubes with integrated caps or seals minimizes the chances of sample contamination and cross-contamination between reactions.
- No Post-PCR Processing: Unlike traditional PCR methods that often require post-PCR processing steps, real-time PCR eliminates the need for additional manipulations after amplification. The results can be obtained directly from the reaction tube, streamlining the workflow and saving time.
- Improved Turnaround Time and Increased Throughput: Real-time PCR enables faster data acquisition, analysis, and interpretation compared to traditional PCR methods. The technique has a shorter turnaround time, allowing for rapid results. Additionally, real-time PCR can be easily scaled up, increasing the throughput and enabling the analysis of a large number of samples simultaneously.
- Multiplexing Capabilities: Real-time PCR offers the ability to perform multiplexing, where multiple target sequences can be amplified and detected simultaneously in a single reaction. This saves time and resources by reducing the number of separate reactions required.
These advantages highlight the efficiency, accuracy, and versatility of real-time PCR, making it a powerful tool in various research fields, clinical diagnostics, and molecular biology applications.
Disadvantages of Real-Time PCR
Real-time PCR (RT-PCR) also has some disadvantages that should be considered. Here are the key disadvantages of real-time PCR:
- Exponential Increase in PCR Product: During real-time PCR, the amplification of the target DNA or RNA occurs exponentially, which means that the product quantity doubles with each cycle. This exponential amplification makes it difficult to accurately determine the size of the amplicon produced.
- Variation with Cycle Number: As the number of PCR cycles increases, there is an inherent variability in the reaction. This can lead to differences in the quantification and amplification of target molecules, resulting in decreased precision and reliability of the assay.
- Limited Simultaneous Reactions: The number of simultaneous reactions that can be performed in real-time PCR is limited, typically up to four reactions. This limitation can restrict the throughput and scalability of the technique when analyzing a large number of samples or targets.
- Spectral Overlap: In fluorescent-based real-time PCR, different fluorophores are used to detect multiple targets. However, some fluorophores may have overlapping emission spectra, making it challenging to accurately distinguish and quantify each target in multiplex assays.
- Risk of False-Negative Results: Real-time PCR may have an increased risk of false-negative results, particularly when detecting new emerging or highly variable pathogens. This can occur if the primers or probes used in the assay do not perfectly match the genetic sequences of the target pathogens, leading to undetected infections.
- Non-Specific Binding: In SYBR Green-based real-time PCR, non-specific binding of the dye to non-target DNA can occur, leading to false-positive signals. This can affect the specificity and accuracy of the assay, requiring careful optimization and validation of the primer design and reaction conditions.
- Amplification of RNA Only: Real-time PCR is primarily designed for the amplification and detection of RNA molecules, especially mRNA. This limits its application to RNA-based analyses and may not be suitable for DNA-specific studies.
- Requirement for Prior Sequence Information: To design specific primers for real-time PCR, prior knowledge of the target RNA or DNA sequence is necessary. This prerequisite may pose challenges when studying novel or unknown targets without available sequence information.
- Sensitivity to Reaction Conditions: Real-time PCR is a temperature and enzyme-dependent technique. Even slight changes in reaction temperature or suboptimal reaction conditions can significantly impact the efficiency and accuracy of the enzyme, leading to decreased amplification efficiency and potentially affecting the reliability of the results.
- Contamination and False Results: Real-time PCR is sensitive to contamination, as even minute amounts of contaminants with primer binding sites can be amplified, resulting in false-positive or false-negative results. Strict laboratory practices and appropriate controls are essential to minimize the risk of contamination.
- Complex and Tedious Process: Performing real-time PCR requires a complex reaction mixture and specialized equipment. It also requires a skilled operator who has expertise in primer design, reaction setup, and data analysis. The process can be time-consuming and demanding, necessitating attention to detail and careful execution.
Considering these disadvantages, it is important to weigh the benefits and limitations of real-time PCR when choosing the appropriate technique for specific applications.
Application of Real-Time PCR
Real-time PCR (RT-PCR) has a wide range of applications in various fields. Here are some key applications of real-time PCR:
- Study Gene Expression: Real-time PCR is widely used to analyze gene expression levels. By amplifying and quantifying mRNA samples, researchers can study the expression patterns of specific genes. It enables the identification and characterization of multidrug-resistant genes in pathogens, aiding in understanding drug resistance mechanisms and designing effective treatments.
- Identification of Unknown Species: RT-PCR plays a crucial role in identifying various microorganisms, including viruses (e.g., HIV, SARS viruses, dengue viruses, HCV), bacteria, fungi, and parasites. By targeting specific rRNA or mRNA sequences, researchers can detect and identify unknown species in diverse samples, such as clinical samples, environmental samples, and food samples.
- Infectious Disease Diagnosis: Real-time PCR is extensively used in clinical laboratories for the diagnosis of infectious diseases. It enables rapid and accurate detection of viral, bacterial, fungal, and parasitic infections. RT-PCR-based tests can identify the presence of pathogenic DNA or RNA in patient samples, aiding in early diagnosis and appropriate treatment selection.
- Gene Insertion and Gene Therapy Study: RT-PCR is employed in gene insertion experiments and gene therapy research. It is used to prepare complementary DNA (cDNA) from eukaryotic mRNA, which lacks introns. This cDNA can be inserted into prokaryotes or used as a template for further genetic manipulations. Real-time PCR is utilized to monitor gene expression levels after insertion and assess the success of gene therapy procedures.
- Study Mutation and Cancer Cells: Real-time PCR allows the detection and quantification of tissue-specific mutant alleles, facilitating the study of genetic mutations associated with diseases. It can identify changes in mRNA sequences, including unique mRNA molecules produced by different types of cancer cells. This enables researchers to investigate genetic alterations and analyze cancer cell behavior.
- Tools of Genetic Engineering and Viral Study: RT-PCR is an essential tool in genetic engineering research. It is used to analyze modified DNAs and their transcribed RNAs, allowing researchers to confirm successful genetic modifications and assess the expression of target genes. Real-time PCR also plays a vital role in viral studies, including viral load quantification, viral genotyping, and monitoring antiviral treatment efficacy.
These applications demonstrate the versatility and significance of real-time PCR in various scientific fields, contributing to advancements in molecular biology, diagnostics, genetic engineering, and disease research.
Difference between One-Step RT-qPCR and Two-Step RT-qPCR
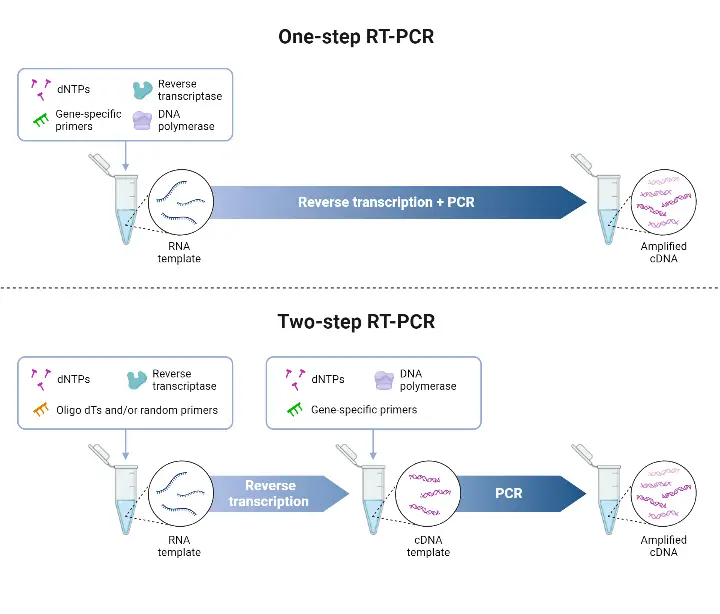
| One-Step RT-qPCR | Two-Step RT-qPCR |
|---|---|
| Single-tube reaction where reverse transcription and PCR amplification occur simultaneously | Two separate reactions: reverse transcription and PCR amplification |
| Reverse transcription and PCR amplification happen in a single thermocycler program | Separate thermocycler programs are used for reverse transcription and PCR amplification |
| Requires a single reaction mixture containing RNA sample, reverse transcriptase, primers, DNA polymerase, and other components | Requires two reaction mixtures: one for reverse transcription with reverse transcriptase, primers, and other components, and another for PCR amplification with DNA polymerase, primers, and other components |
| Offers convenience and simplicity with fewer pipetting steps and reduced hands-on time | Involves more pipetting steps and longer hands-on time due to separate reactions |
| Suitable for high-throughput applications and limited starting material | Preferred for applications where flexibility in selecting different enzymes for reverse transcription and PCR is required |
| Ideal for gene expression analysis and detection of RNA targets in a single step | Allows for storage of cDNA for later use and analysis of multiple targets using the same RNA sample |
| Higher risk of contamination due to the open-tube nature of the reaction | Lower risk of contamination as reverse transcription and PCR amplification are performed in separate tubes |
| May result in higher chances of non-specific binding and primer-dimer formation | Lower chances of non-specific binding and primer-dimer formation |
| Provides accurate and reliable results with high efficiency | Offers high efficiency, accuracy, and reliability in detecting larger templates per reaction mixture |
FAQ
What is real-time PCR?
Real-time PCR, also known as quantitative PCR (qPCR), is a molecular biology technique used to amplify and quantify specific DNA or RNA sequences in real time during the PCR process.
How does real-time PCR differ from conventional PCR?
In conventional PCR, amplification results are analyzed after the reaction is completed, whereas real-time PCR allows for continuous monitoring of the amplification process and quantification of the target DNA or RNA during the reaction.
What are the key components required for real-time PCR?
Real-time PCR requires a thermal cycler instrument with the ability to measure fluorescence signals, specific primers for the target sequence, a DNA polymerase enzyme, fluorescent probes or dyes, and a sample containing the target DNA or RNA.
What are the advantages of real-time PCR?
Real-time PCR offers several advantages, including rapid amplification and quantification of target sequences, high sensitivity and specificity, broad dynamic range for quantification, and the ability to analyze multiple targets simultaneously.
How is real-time PCR used in gene expression studies?
Real-time PCR is widely used to study gene expression by quantifying the amount of mRNA present in a sample. It allows researchers to measure changes in gene expression levels under different conditions or in different tissues.
What are the different fluorescence markers used in real-time PCR?
Commonly used fluorescence markers in real-time PCR include SYBR Green, TaqMan probes, molecular beacons, and hybridization probes. These markers bind to the amplified DNA or RNA and emit fluorescent signals that are measured during the reaction.
How is data analysis performed in real-time PCR?
Data analysis in real-time PCR involves determining the cycle threshold (Ct) value, which represents the cycle number at which the fluorescence signal crosses a predefined threshold. The Ct value is used to quantify the initial amount of the target DNA or RNA in the sample.
Can real-time PCR be used for pathogen detection?
Yes, real-time PCR is commonly used for pathogen detection, including viruses, bacteria, and parasites. It offers high sensitivity and specificity, allowing for the rapid and accurate identification of infectious agents in clinical samples.
What is the difference between absolute and relative quantification in real-time PCR?
Absolute quantification in real-time PCR determines the exact amount of target DNA or RNA present in a sample by comparing the Ct values to a standard curve generated using known concentrations. Relative quantification compares the expression levels of the target gene to a reference gene or control sample without determining the absolute copy number.
Can real-time PCR detect and quantify genetic mutations?
Yes, real-time PCR can be used to detect and quantify genetic mutations by designing specific primers and probes that target the mutated sequence. This allows researchers to identify and quantify the presence of mutant alleles in a sample.
References
- International Journal of Multidisciplinary and Current Research Detection of Water-borne Pathogenic Bacteria: Where Molecular Methods Rule – Scientific Figure on ResearchGate. Available from: https://www.researchgate.net/figure/Advantages-and-Limitations-of-Real-Time-PCR_tbl2_292318147 [accessed 27 May, 2021]
- https://international.neb.com/protocols/2014/08/15/one-step-rt-pcr-protocols-e5315
- https://ocw.mit.edu/courses/biology/7-16-experimental-molecular-biology-biotechnology-ii-spring-2005/labs/rt_pcr_2step.pdf
- https://microbenotes.com/real-time-pcr-principle-process-markers-advantages-applications/
- https://www.thermofisher.com/in/en/home/brands/invitrogen/molecular-biology-technologies/spotlight-articles/onestep-vs-twostep-rtpcr.html
- https://himedialabs.com/TD/HTBM032.pdf
