Table of Contents
What is Small nuclear RNA (snRNA)?
- Small nuclear RNA (snRNA) is a small RNA with an average length of 150 nucleotides.
- A variety of noncoding RNAs are encoded by eukaryotic genomes, and snRNA is a kind of highly abundant, nucleus-localized RNA with essential roles in intron splicing and other RNA processing.
- In order to mediate the process of transcript splicing, snRNA typically exists as ribonucleoprotein particles (snRNPs) with other proteins that create a huge particulate complex (spliceosome) bound to the unspliced primary RNA transcripts.
- In addition to splicing, snRNPs function in nuclear maturation of initial transcripts in mRNAs, gene expression regulation, splice donor in non-canonical systems, and 3′-end processing of replication-dependent histone mRNAs, according to new data.
- Small nuclear ribonucleic acid (snRNA), often known as U-RNA, is a category of small RNA molecules present in the nucleus of eukaryotic cells.
- Intronless, non-polyadenylated, non-coding transcripts with nuclear function.
- Small nuclear ribonucleoprotein complexes are always composed of SnRNA and a set of particular proteins (snRNP often pronounced “snurps”)
- Each snRNP particle consists of snRNA and multiple snRNP-specific proteins.
Types of Small nuclear RNA (snRNA)
The snRNAs can be divided into two classes on the basis of common sequence features and protein cofactors.
1. Sm-class RNAS
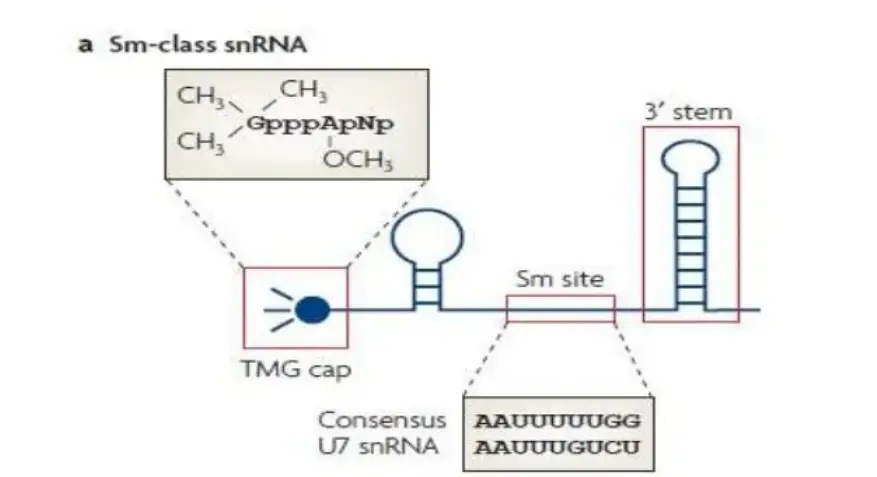
- This is characterised by a 5′-trimethylguanosine cap, a 3′ stem-loop, and a binding site for a heteroheptameric ring-shaped group of seven Sm proteins (the Sm site).
- It includes U1, U2, U4,U4atc, U5, U7, U11, and U12 teams.
- The RNA polymerase II (Pol II) used to transcribe Sm-class genes is functionally comparable to the Pol II used to transcribe mammalian protein-coding genes.
- Sm-class snRNAs are exported from the nucleus to undergo maturation in the cytoplasm.
2. Lsm-class RNAs
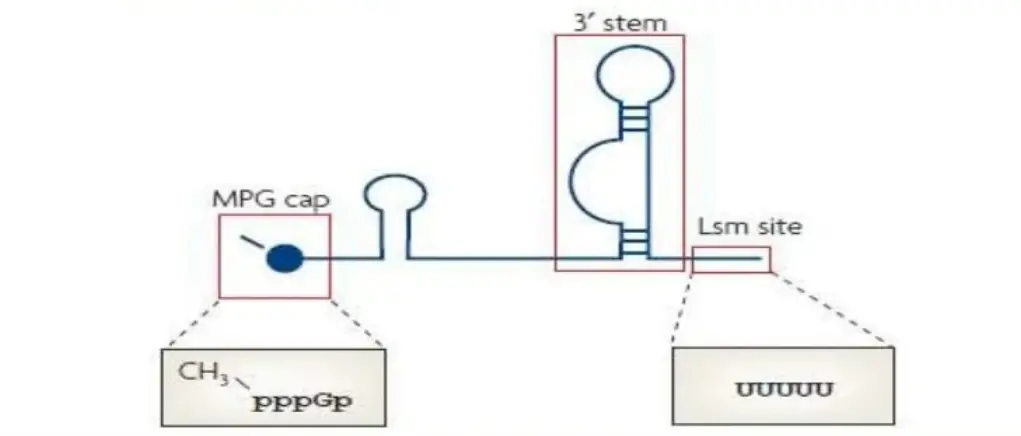
- This class possesses a monomethyl phosphate cap and a 3′ stem-loop, the latter of which terminates in a stretch of uridines that serve as the binding site for an unique heteroheptameric ring of Lsm proteins.
- It contains U6 and Ubatac
- The Lsm-class snRNA genes (U6 and Ubatac) are transcribed by Pol III using external Promoters that are specialised for this purpose.
- snRNAs of the Lsm class never leave the nucleus.
Structure of snRNA
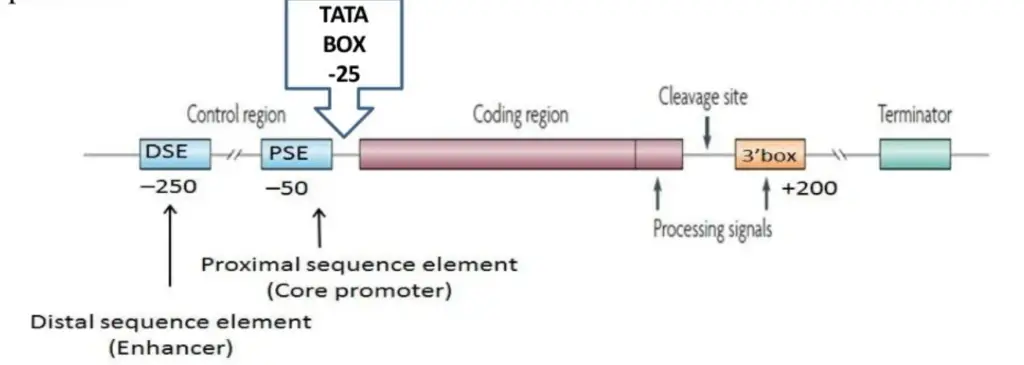
- Distal sequence element (DSE) works as an enhancer.
- Required connection between the 3′ box RNA processing element and the promoter of the snRNA gene type.
- In addition to a DSE and PSE, the pol III-transcribed genes have a TATA box at the -25 location.
- The core promoter is an important snRNA gene-specific proximal sequence element (PSE).
Steps of snRNA gene transcription
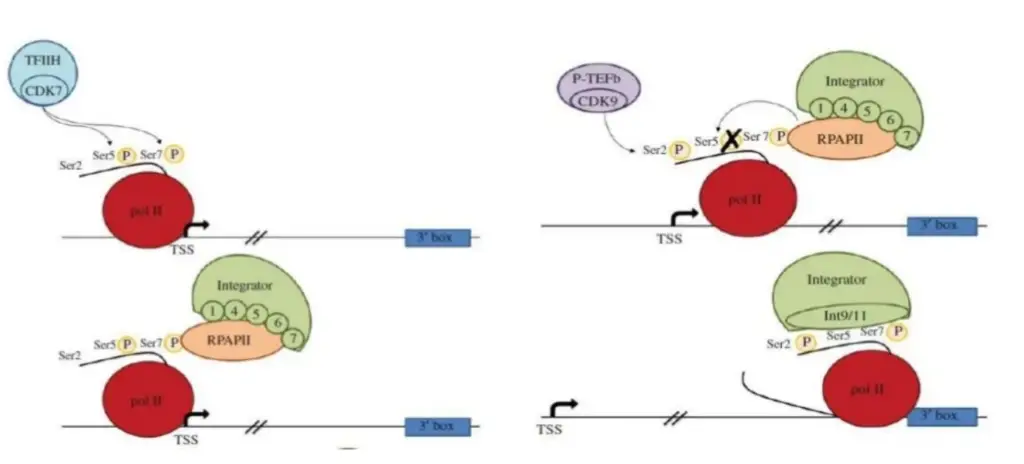
phosphorylation of the CTD of Pol II during snRNA gene transcription. Initial phosphorylation of Ser5 and Ser7 by the cyclin-dependent kinase (CDK)7 subunit of TFIIH. Ser7P interacts with RPAP2. The Integrator subunits Int1, Int4, Int5, Int6, and Int7 are recruited by RPAP2. Unknown mechanism recruits positive-transcription elongation factor b (P-TEFb) after RPAP2 dephosphorylates Ser5P. CDK9, a subunit of P-TEFb, phosphorylates Ser2. The double phosphorylation of Ser2 and Ser7 recruits the RNA processing enzyme Int9/11.
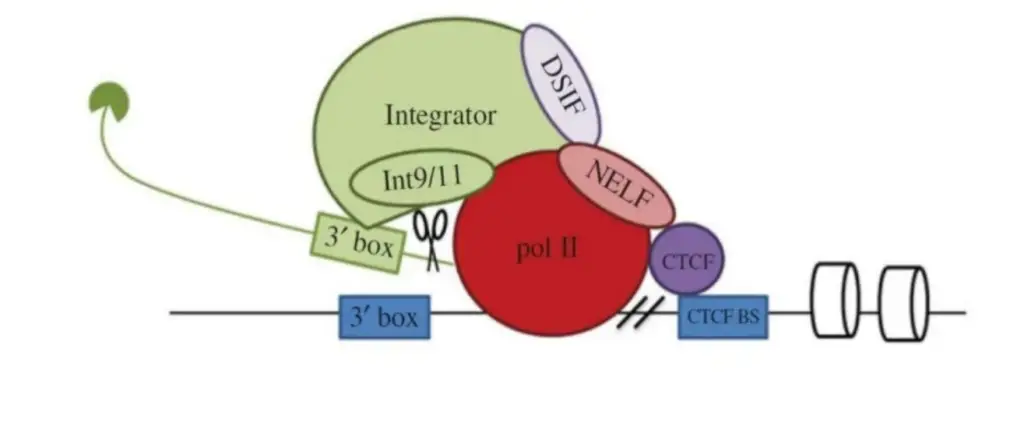
Transcription termination of the snRNA gene
Model for the termination of U2 snRNA gene transcription. The snRNA transcript is shown in green with a cap at the 5′ end, whereas nucleosomes are depicted as barrels. Pol Il continues transcription after recognising the 3′ box, whereas Integrator processes the newly synthesised RNA. CTCF identifies the CTCF binding site upstream of the 3′ box and regulates nucleosome occupancy. Negative elongation factor (NELF) is recruited by DRB sensitivity-inducing factor (DSIF) and CTCF at the end of the transcription unit and causes transcription termination
References
- Hari, R., & Parthasarathy, S. (2018). Prediction of Coding and Non-Coding RNA. Reference Module in Life Sciences. doi:10.1016/b978-0-12-809633-8.20099-x
- Buratti, Emanuele. (2013). Small Nuclear RNA. 10.1016/B978-0-12-374984-0.01437-6.
- https://www.frontiersin.org/articles/10.3389/fgene.2021.652129/full
- https://mcmanuslab.ucsf.edu/rna/small-nuclear-rna
- https://www.genscript.com/biology-glossary/2739/small-nuclear-rna-snrna
- https://www.slideshare.net/RushilMandlik/small-nuclear-rna
- https://www.ebi.ac.uk/chebi/searchId.do;?chebiId=CHEBI:74035


