Table of Contents
What is Spliceosome?
- Spliceosomes are large, multi megadalton ribonucleoprotein (RNP) complexes found in eukaryotic nuclei.
- They assemble on RNA polymerase II transcripts, from which they remove introns and splice exons.
- This so-called pre-messenger RNA (pre-mRNA) splicing is a crucial stage in the synthesis of eukaryotic messenger RNA (mRNA).
- Each human cell includes approximately 100,000 spliceosomes, which are responsible for the removal of more than 200,000 distinct intron sequences.
- Post-transcriptional processing, which is required for the creation of functional mRNA in eukaryotes, includes splicing.
- Splicing takes place in the nucleus. In this process, the non-coding introns of pre-mRNA are eliminated, and the coding exons are brought together.
- Occasionally, RNA catalyses its own splicing. It is known as a ribozyme.
- Self-splicing does not require spliceosomes.
- Spliceosome is a massive protein complex composed of small nuclear ribonucleoproteins (snRNPs, sometimes known as “snurps”).
- The spliceosome complexes are multi-megadalton in size. In humans, around 300 distinct types of proteins assemble to create the spliceosome.
- Each cell has approximately one hundred thousand spliceosomes that delete diverse intron sequences.
- Some of the spliceosome proteins are specialised to RNA, while others drive the process to ensure its correctness.
- Small nuclear RNAs (snRNAs) generate small nuclear riboprotein complexes by binding to other proteins (snRNPs).
- Uridine is abundant in the snRNAs that make up the spliceosome complex. They are designated as U1, U2, U4, U5, and so on.
- Two types of spliceosomes exist:
- Major Spliceosomes – They are the reason why 99.5% of introns are taken out. They are made up of the snRNPs U1, U2, U4, U5, and U6.
- Minor Spliceosomes – They are the reason why 0.5% of introns are taken out. They are made up of snRNPs with the sequences U11, U12, U4atac, U5, and U6atac.
Properties of Spliceosome
- Proteins and RNA molecules are both found in spliceosomes.
- There are just about a hundred spliceosomal proteins in yeast, whereas in humans there are about three hundred.
- Many of these proteins are RNA-recognition experts, while others are nucleotide-binding proteins (NTPases) whose job it is to propel the process along and guarantee its accuracy.
- Multiple additional proteins attach to snRNAs in a stable fashion to generate tiny nuclear RNPs (snRNPs, pronounced’snurps’).
- U1, U2, U4, U6, and U5 snRNPs (called for the U snRNA(s) they contain) are organised into major spliceosomes.
- U11, U12, U4atac, U5, and U6atac snRNPs come together to form minor spliceosomes.
Structure and composition of Spliceosome
- Five snRNAs and various protein components work together to form a spliceosome.
- RNA-protein complexes, or snRNPs, are formed when these short RNAs bind to their respective protein components (small nuclear ribonucleoproteins, pronounced “snurps”).
- The main spliceosome is comprised of the snRNAs U1, U2, U4, U5, and U6, which were given these names because they are highly enriched in the uridine and are involved in multiple RNA-RNA and RNA-protein interactions.
- Each exon:intron junction on a pre-mRNA (also called heterogeneous nuclear RNA, hn-RNA) is the site of spliceosome assembly.
- Spliceosome assembly makes use of particular sequence components found in the introns of pre-mRNA. Included in this category are the 5′ end splice site, 3′ end splice site, branch point sequence, and polypyrimidine tract.
- The spliceosome acts as an enzyme complex responsible for splicing out introns and joining together adjacent exons.
- It is common for introns to have a GU nucleotide sequence at the 5′ end splice site and an AG sequence at the 3′ end splice site.
- To further characterise the 3′ splice site, a variable length of polypyrimidines called the polypyrimidine tract (PPT) can be inserted. The PPT serves the dual purpose of recruiting factors to the 3′ splice site and, potentially, recruiting factors to the branch point sequence (BPS).
- The first splicing process necessitates the presence of a conserved adenosine, which is found in the BPS.
- Since zinc is crucial to the splicing process, the presence of zinc-binding motifs in many proteins is not surprising.
- In 2016, scientists announced the first molecular-resolution reconstruction of the U4/U6/U5 tri-snRNP complex.
- Shi et al. have made considerable use of cryo-EM to determine the near-/atomic structure of the human and yeast spliceosomes.
- Spp42, a component of U5 snRNP, is shown to anchor the catalytic centre of the spliceosome and to form a central scaffold in a near-atomic-resolution model of the yeast spliceosome.
- Step II component Slu7 adopts an expanded structure in the human spliceosome, ready to select the 3′-splice site, as shown by the structure at the atomic level.
- The human complex retains the same five metals (designated as Mg2+) as the yeast complex.
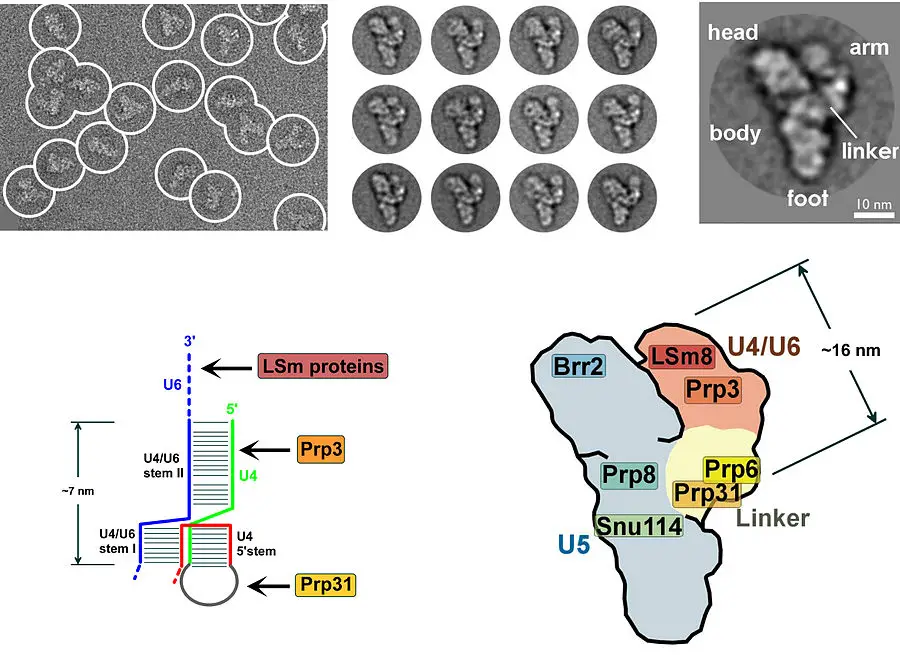
Components of Spliceosome
In addition to the pre-mRNA, spliceosomes include many other RNAs and proteins. Small nuclear ribonucleoproteins (snRNPs, pronounced “snurps”) are a type of RNA-protein complex that includes a small nuclear RNA (snRNA) bound to a protein. Gel electrophoresis enables the snRNAs to be separated into the species U1, U2, U4, U5, and U6. These five RNAs all assemble into the spliceosome and perform essential functions during the splicing process.
U1 snRNP
- A portion of U1 snRNA contains a sequence that is nearly completely complementary to consensus sequences found at both 5′ and 3′ splice sites.
- The U1 snRNA formed a base pair with these splice sites, bringing them together for the splicing process.
- Splicing relies on the base pairing between U1 snRNA and the 5′ splice site.
U6 snRNP
- The U6 snRNA mediates the association of the U6 snRNP with the 5′ end of the intron.
- This pairing happens in the early stages of splicing, before the lariat intermediate is formed, and it continues after this initial stage of splicing is complete.
- Splicing cannot occur without U6 first associating with the splicing substrate.
- During splicing, U6 also forms associations with U2.
U2 snRNP
- At the splicing branchpoint, the U2 snRNA forms base pairs with a conserved sequence.
- This pairing of bases is required for splicing.
- U2 also forms essential base pairs with U6 to form a region known as helix I, which appears to aid in the orientation of these snRNPs during splicing.
- Additionally, the 5′-end of U2 interacts with the 3′-end of U6 to produce helix II, which is essential for splicing in mammalian cells but not yeast cells.
U5 snRNP
- The U5 snRNA binds with the last nucleotide of one exon and the first nucleotide of the subsequent exon.
- This likely aligns the two exons in preparation for splicing.
U4 snRNP
- U4 forms a base-pair with U6, and its job appears to be to bind U6 until it is required for the splicing reaction.
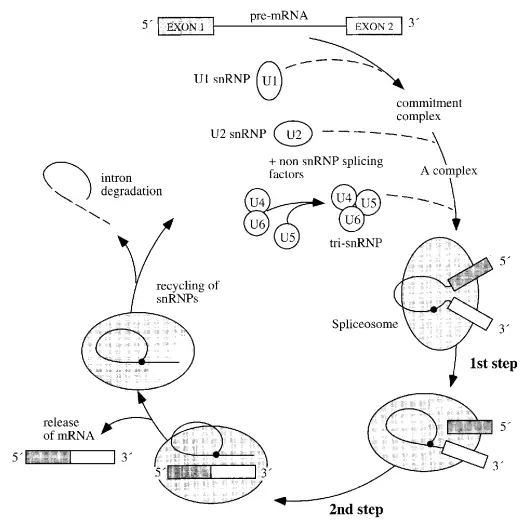
Spliceosome Assembly or The Spliceosome Cycle
Spliceosome assembly, splicing activity, and disassembly comprise the spliceosome cycle. A commitment complex is formed upon the binding of U1 to the splicing substrate. The following snRNP to join the complex is U2, then the others. The binding of U2 is dependent on ATP. U6 dissociates from U4 and displaces U1 at the 5′-splice location upon dissociation. This ATP-dependent phase activates the spliceosome and permits the release of U1 and U4.
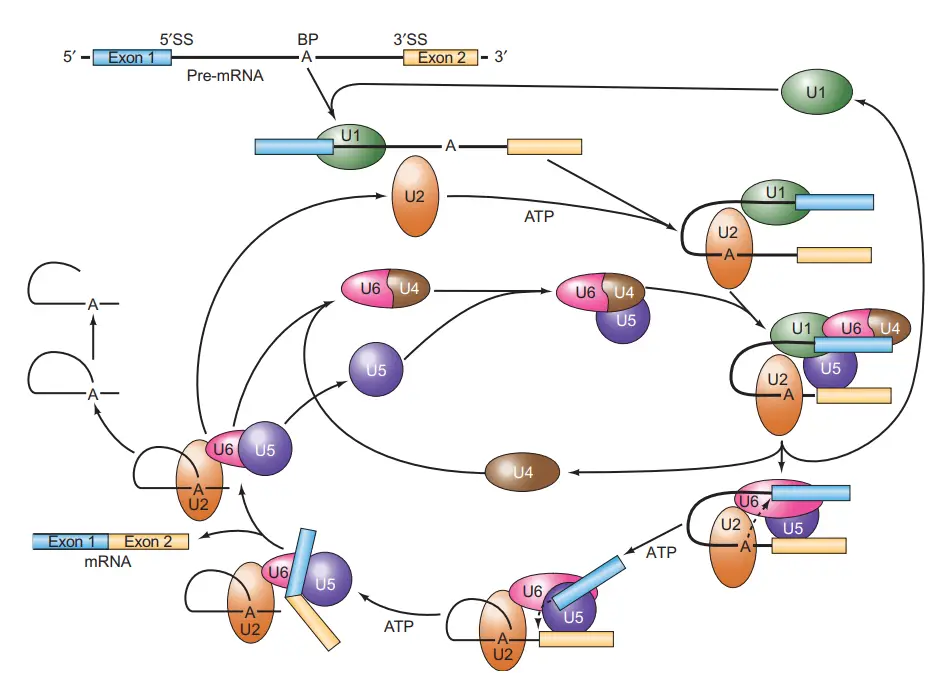
- Composed of splicing substrate, U1, and maybe other chemicals, the first complex to form is known as the commitment complex (CC).
- The commitment complex, as its name suggests, is committed to splicing out the intron at which it assembles.
- Following this, U2 joins ATP to form the A complex.
- U4–U6 and U5 then combine to produce the B1 complex. U4 dissociates from U6 to permit: (1) U6 to displace U1 from the 5′-splice site in an ATP-dependent process that activates the spliceosome; (2) U1 and U4 to exit the spliceosome; and (3) U6 to base-pair with U2.
- The spliceosome that has been triggered is also known as the B2 complex.
- Then, ATP provides the energy for the first splicing step, which separates the two exons and generates the lariat splicing intermediate, both of which are included inside the C1 complex.
- Using energy from a second molecule of ATP, the C2 complex performs the second splicing process, connecting the two exons and eliminating the lariat-shaped intron.
- The mature, spliced mRNA then leaves the I complex, leaving the intron linked to the complex.
- The I complex dissociates into its constituent snRNPs, which can be recycled into another splicing complex, and the debranched and degraded lariat intermediate.
Mechanism of Splicing
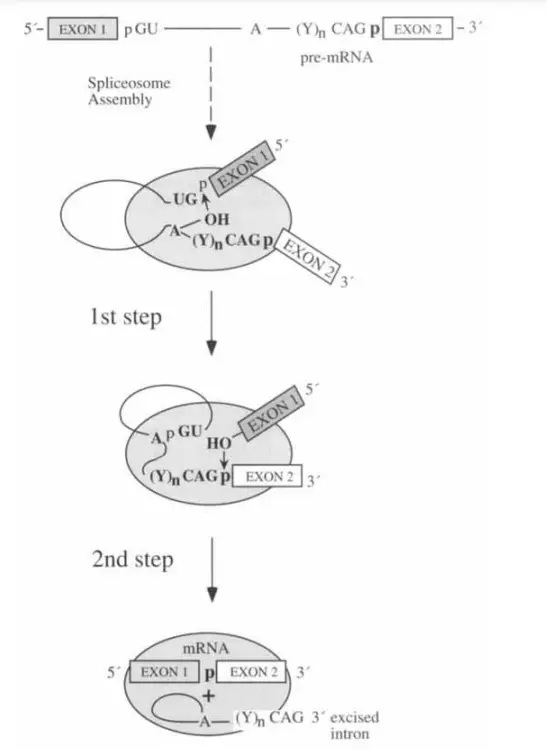
- Splicing is the process by which introns in pre-mRNA or hnRNA are excised and exons are joined together.
- When hnRNA is being processed, the spliceosome is assembled at the exon-intron junction.
- The U1 snRNP may identify the 5′-end of an intron.
- A specific location is located close to the 3′-end of an intron that is recognised by the U2 snRNP. Rearrangement and assembly of the remaining parts follows.
- There are two phases to the splicing:
- Cleavage at the intron’s 5′ splice point is the initial process.
- The intron’s 3′ splice site is cleaved off and the exons are linked together in the second phase. These two responses occur simultaneously.
- Once the intron has been excised, the spliceosome that was encasing it detaches and the intron is damaged.
- Since introns are present in most human genes, spliceosomes play a role in how those genes are expressed.
- This phenomenon, known as alternative splicing, occurs in some genes and results in the production of several RNAs that encode for distinct proteins.
- Mutation and genetic disorders may result from faulty splicing of genes or from spliceosomes that are not properly regulated.
- A protein’s structure and abundance can be drastically altered by even a single point mutation.
snRNP Structure
- The same seven Sm proteins are present in all snRNPs.
- Antibodies directed against these proteins are frequently found in people with autoimmune disorders like systemic lupus erythematosus.
- The Sm proteins were really named after Stephanie Smith, the SLE patient in whose blood they were first found.
- There is a shared binding site between the Sm proteins. snRNAs with the sequence (AAUUUGUGG).
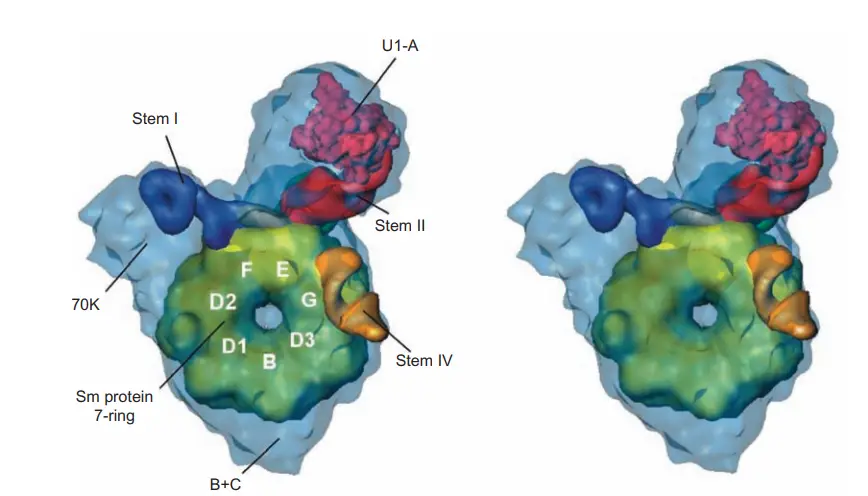
- Each snRNP contains its own unique group of proteins in addition to the Sm proteins. The three individual proteins that make up U1 snRNP have Mr’s of 52, 31, and 17.5 kD, respectively; these are designated 70K, A, and C.
- By employing single-particle electron cryomicroscopy, Holger Stark and coworkers were able to determine the structure of the U1 snRNP to a resolution of 10 angstroms. As can be seen in the structure, the Sm proteins pack together to form a doughnut with a hole in the middle, like a flattened funnel.
- A and 70K, the two biggest U1-specific proteins, are linked to the Sm “doughnut” and bind to stem loop motifs in U1 snRNA.
- We were able to determine which protrusions correspond to the 70K and A proteins by using electron microscopy to look for missing protrusions in negatively stained samples of U1 snRNPs that were depleted of these proteins.
- The Sm site RNA is in a single-stranded area, so it might travel through the “doughnut” gap.
- Previous x-ray crystallographic investigations on Sm protein subassemblies had actually predicted a ring-shaped structure with a hole bordered with basic amino acid side chains.
- Such a hole’s fundamental nature would promote binding to the Sm site in the U1 snRNA.
Minor spliceosome
- An unusual sort of spliceosomal introns (U12-type) are removed (spliced) from eukaryotic messenger RNAs by a ribonucleoprotein complex known as the minor spliceosome. This complex is found in plants, insects, vertebrates, and some fungi (Rhizopus oryzae).
- In contrast to U2-dependent canonical splicing, this is referred to as noncanonical splicing. U12-type introns account for fewer than 1% of all introns in human cells.
- On the other hand, they show up in genes involved in crucial biological processes.
- In the middle of the 1990s, a unique form of intron was found in metazoans (animals with distinct organs).
- When compared to their counterparts in the major introns, the 5′-splice sites and branchpoint sequences in these variant introns are substantially conserved yet significantly dissimilar.
- This discovery prompted the question of how transcripts of genes with variable introns could be spliced if the intron sequences did not match those of any of the known snRNAs. The answer is that minor spliceosomes are present in metazoan cells and are composed of the snRNAs U11 (which acts like U1), U12 (which acts like U2), U4atac (which acts like U4), and U6atac (which acts like U6).
- Both the major and minor spliceosomes rely on U5 snRNA.
- This method of alternative splicing is a check on the significance of base pairing between snRNAs and critical locations in pre-mRNAs.
- The branchpoint in the alternative pre-mRNAs is base-paired with U12 snRNA, while the 5′-splice site is base-paired with U11.
- Not only that, but U4atac and U6atac can base pair with each other just like U4 and U6 can.
- Do we know anything about the proteins that assemble into snRNPs from the minor snRNAs? The first thing to note is that, in addition to the separate U11 and U12 snRNPs, there is also an U11/U12 snRNP.
- To a lesser extent than with the major snRNPs, U11 and U12 snRNPs have their own set of related proteins.
- The seven Sm proteins are present in all of the main snRNPs and represent an example of a protein family that is widely distributed.
- Separation of the major and minor spliceosomes: We now know that the major spliceosome is located in the nucleus, while the minor spliceosome is primarily located in the cytoplasm.
- Some transcripts have introns that are recognised by the major spliceosome, whereas others contain introns that are recognised by the minor spliceosome.
- Following removal of the major spliceosomal introns in the nucleus, the partially spliced pre-mRNA exits the nucleus and its minor introns are eliminated in the cytoplasm. Not much is known about the physiological significance of this division of labour.
Spliceosomes associated with many diseases
- Missplicing of a single gene or dysregulation of the spliceosome as a whole are both major contributors to many human illnesses.
- Around one-third of all human genetic illnesses result from a mutation that changes the splicing of just one gene.
- These changes can be as simple as the addition or removal of a single splice site (as seen in – or -thalassemia) or as complex as the alteration of the ratio of alternative spliced exons (e.g., frontotemporal dementia driven by tau mis-splicing).
- Sometimes an mRNA isoform is produced as a result of incorrect splicing that is then rapidly degraded.
- As a result, single point mutations can drastically alter protein structure and protein abundance through their effects on splicing.
- Mutations in the splicesomal proteins themselves cause other illnesses by altering the splicing of many transcripts. It has been established that mutations in a number of essential spliceosomal proteins (such as Prp8, Prp3, Prp31, and Brr2) are the direct cause of autosomal dominant retinitis pigmentosa.
- Chronic lymphocytic leukaemia and myelodysplasia are often linked to mutations in splicing factor 3B subunit 1 (SF3B1) and U2 auxiliary factor 35 (U2AF35).
- Misregulation of splicing factor levels has also been linked to other types of cancer.
- As a result, the spliceosome is now being studied as a potential therapeutic target for cancer.
How do spliceosomes affect gene expression?
- Splicing is a crucial part of gene expression since nearly all human protein-coding genes contain introns (usually 9 or 10, but some have more than 100!).
- Alternative splicing, which facilitates the production of several mRNA variants from a single DNA gene, has recently been found to affect over 95% of human genes, as discovered by high-throughput sequencing.
- Alternative spliced mRNAs considerably increase biological complexity by encoding distinct protein isoforms or storing diverse regulatory sequences in their untranslated regions.
- Besides the obvious effects of intron removal, the process of splicing itself can have profound effects on gene expression.
- Splicing can modify mRNA’s subcellular localization, translation efficiency, and decay kinetics by permanently depositing on exons proteins that travel with mRNPs to the cytoplasm (such as the exon junction complex, EJC).
- In particular, the relative position of the EJC to the stop codon on the mRNA is a critical mediator of protein abundance in the cell.
References
- Lamond AI. The spliceosome. Bioessays. 1993 Sep;15(9):595-603. doi: 10.1002/bies.950150905. PMID: 8240312.
- Zhang, X., Yan, C., Hang, J., Finci, L. I., Lei, J., & Shi, Y. (2017). An Atomic Structure of the Human Spliceosome. Cell, 169(5), 918–929.e14. doi:10.1016/j.cell.2017.04.033
- Will CL, Lührmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol. 2011 Jul 1;3(7):a003707. doi: 10.1101/cshperspect.a003707. PMID: 21441581; PMCID: PMC3119917.
- https://www.sciencedirect.com/topics/medicine-and-dentistry/spliceosome
- https://cshperspectives.cshlp.org/content/3/7/a003707
- https://www.cell.com/current-biology/pdf/S0960-9822(14)01553-X.pdf
- https://cshperspectives.cshlp.org/content/3/7/a003707/F2.expansion.htm
