Table of Contents
What is Wobble Hypothesis?
The Wobble Hypothesis, proposed by Francis Crick in 1966, provides an explanation for the degeneracy of the genetic code. Degeneracy refers to the fact that multiple codons can code for the same amino acid. According to the Wobble Hypothesis, the precise pairing between the bases of the codon and the anticodon of tRNA occurs only for the first two bases of the codon. However, the pairing between the third base of the codon and the anticodon can exhibit some flexibility or “wobble.”
In other words, the third base of the codon and the anticodon can sway or move unsteadily, allowing for non-standard base pairing. This phenomenon enables a single tRNA molecule to recognize and bind to multiple codons, despite differences in their third base. As a result, although there are 61 codons that code for amino acids, the number of tRNA molecules is significantly lower (around 40) due to wobbling.
Wobble base pairs play a crucial role in RNA secondary structure and are critical for accurate translation of the genetic code. The four main wobble base pairs are guanine-uracil (G-U), hypoxanthine-uracil (I-U), hypoxanthine-adenine (I-A), and hypoxanthine-cytosine (I-C). These wobble base pairs exhibit thermodynamic stability comparable to that of Watson-Crick base pairs.
In the genetic code, there are 64 possible codons, out of which three are stop codons that terminate translation. If canonical Watson-Crick base pairing were required for each codon, it would necessitate 61 different types of tRNA molecules. However, most organisms have fewer than 45 types of tRNA. The Wobble Hypothesis explains how some tRNA molecules can recognize multiple synonymous codons, which encode the same amino acid.
Crick proposed that the 5′ base on the anticodon, which binds to the 3′ base on the mRNA codon, is more flexible spatially compared to the other two bases. This flexibility allows for non-standard base pairing and small conformational adjustments, resulting in the overall pairing geometry of tRNA anticodons.
Overall, the Wobble Hypothesis provides a mechanism to account for the degeneracy of the genetic code and the ability of a limited number of tRNA molecules to recognize and bind to multiple codons. The wobbling of the third base allows for greater flexibility in genetic coding while maintaining the accuracy of translation.
Definition of Wobble Hypothesis
The Wobble Hypothesis proposes that the third base of a codon and the anticodon of tRNA can exhibit flexibility or “wobble” in their base pairing, allowing a single tRNA molecule to recognize and bind to multiple codons, contributing to the degeneracy of the genetic code.
The Wobble Hypothesis
The Wobble Hypothesis proposes that the base at the 5′ end of the anticodon in tRNA is not as constrained spatially as the other two bases. This flexibility allows it to form hydrogen bonds with multiple bases located at the 3′ end of a codon in mRNA. The wobble hypothesis states that:
- The first two bases of the codon and the corresponding bases in the anticodon form normal Watson-Crick base pairs.
- The third position in the codon follows less strict base-pairing rules, leading to non-canonical pairing or “wobble.”
- The relaxed base-pairing requirement at the third position enables a single tRNA molecule to pair with more than one mRNA triplet.
- The specific rules for wobble pairing are: U in the first position of the anticodon can recognize A or G in the codon, G can recognize U or C, and I (inosine) can recognize U, C, or A.
- These characteristics led Francis Crick to propose the wobble hypothesis, which explains the flexible base-pairing interactions observed in the genetic code.
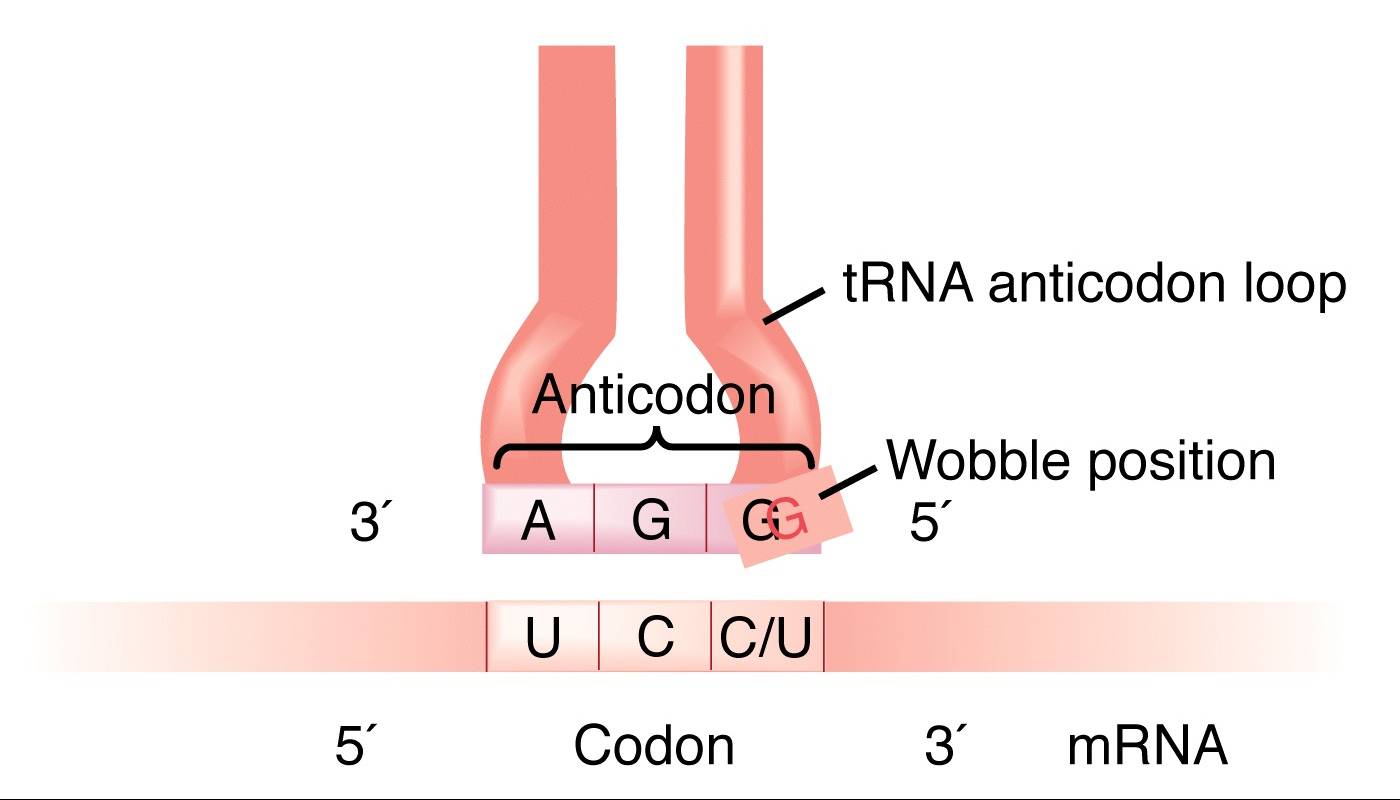
The coding specificity of the genetic code primarily depends on the first two bases of the codon, which form strong Watson-Crick base pairs with the anticodon of tRNA. The first nucleotide in the anticodon determines how many nucleotides the tRNA can distinguish when reading the codon in the 5′ to 3′ direction.
If the first nucleotide in the anticodon is C or A, the pairing is specific, and only one specific codon can be recognized by that tRNA. However, if the first nucleotide is U or G, the pairing is less specific, allowing interchangeability between two bases in the codon. In the case of inosine as the first nucleotide, it exhibits true wobble properties, enabling it to pair with any of three bases in the original codon.
Due to the specificity of the first two nucleotides in the codon, if an amino acid is coded for by multiple anticodons and those anticodons differ in the second or third position (first or second position in the codon), a different tRNA is required for each anticodon.
To satisfy all possible codons (61 excluding three stop codons), a minimum of 32 tRNA molecules is required. This includes 31 tRNAs for the amino acids and one for the initiation codon.
The Wobble Hypothesis provides a key understanding of the flexibility and degeneracy of the genetic code, allowing a limited number of tRNA molecules to recognize and bind to multiple codons while maintaining the accuracy of protein translation.
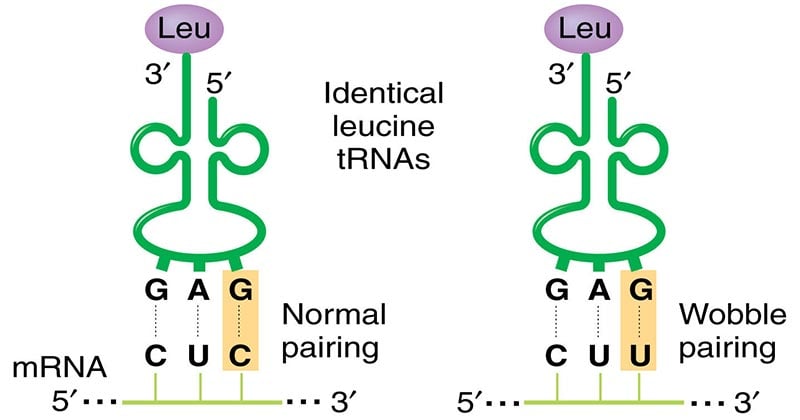
Wobble base pairs
Wobble base pairs are specific pairings between nucleotides in RNA molecules that deviate from the standard Watson-Crick base pair rules. They play a crucial role in RNA structure and translation. Here are some key points about wobble base pairs:
- Wobble base pairs involve non-standard pairings between nucleotides in specific positions.
- The four main wobble base pairs identified in RNA are guanine-uracil (G-U), hypoxanthine-uracil (I-U), hypoxanthine-adenine (I-A), and hypoxanthine-cytosine (I-C).
- The use of “I” for hypoxanthine maintains consistency in nucleic acid nomenclature since hypoxanthine is the nucleobase of inosine.
- Among the wobble base pairs, inosine exhibits the most significant characteristics. If inosine is present as the first nucleotide in the anticodon of tRNA, it can pair with any of three bases (adenine, cytosine, or uracil) in the corresponding codon on mRNA.
- Inosine’s ability to wobble allows a single tRNA molecule with an inosine-containing anticodon to recognize multiple codons, expanding the flexibility of the genetic code.
Wobble base pairs introduce a level of versatility in RNA interactions, particularly during translation. They enable a reduced number of tRNA molecules to recognize multiple codons, compensating for the degeneracy of the genetic code. The wobble hypothesis, which explains these non-standard pairings, provides insights into the efficiency and accuracy of protein synthesis.
Short Exaplanation of The Wobble Hypothesis
Let’s dive into the Wobble Hypothesis step by step.
The Wobble Hypothesis is a concept that explains how the genetic code, stored in the form of nucleotides, is translated into proteins by the ribosome. To understand this hypothesis, we need to know a little bit about codons and anticodons.
Codons are sequences of three nucleotides in mRNA, and each codon corresponds to a specific amino acid or a stop signal. On the other hand, anticodons are sequences of three nucleotides in tRNA that bind to the codon during protein synthesis.
According to the Wobble Hypothesis, the base at the 5′ end of the anticodon is not as restricted in its pairing as the other two bases. This means that the first two bases of the codon and anticodon form normal hydrogen bond pairs, following the usual base-pairing rules (A with U, G with C). However, at the third position of the codon, the rules are more relaxed, and non-canonical pairing can occur.
In other words, the third base of the codon and the first base of the anticodon can form “wobble” pairs that do not strictly follow the A-U and G-C base-pairing rules. This flexibility allows the anticodon of a single tRNA molecule to recognize and bind to more than one codon with different nucleotide sequences at the third position.
To give you some examples of the wobble pairing rules:
- If the first base of the anticodon is U, it can recognize codons with A or G as the third base.
- If the first base of the anticodon is G, it can recognize codons with U or C as the third base.
- If the first base of the anticodon is I (Inosine), it can recognize codons with U, C, or A as the third base.
By allowing this “wobble” or relaxed base-pairing, the cell can minimize the number of tRNA molecules needed for protein synthesis. It provides flexibility and efficiency in translating the genetic code.
So, in summary, the Wobble Hypothesis proposes that the third base of the codon and the first base of the anticodon can form non-standard base pairs, leading to a more flexible set of base-pairing rules at the third position of the codon. This flexibility allows a single tRNA molecule to recognize and bind to multiple codons with different nucleotide sequences at the third position, optimizing protein synthesis.
Importance of the Wobble Hypothesis
The wobble hypothesis holds significant importance in understanding the efficiency and accuracy of protein synthesis. Here are key points highlighting the importance of the wobble hypothesis:
- Broad specificity with limited tRNAs: Our bodies possess a limited number of tRNA molecules. The wobble hypothesis allows a single tRNA to recognize multiple codons due to non-standard pairings at the wobble position. This broad specificity enables efficient translation with a smaller set of tRNAs.
- Facilitation of biological functions: Wobble base pairs have been extensively studied in organisms such as Escherichia coli (E. coli), demonstrating their role in various biological processes. They contribute to the accuracy of translation and protein synthesis.
- Comparable thermodynamic stability: Wobble base pairs exhibit thermodynamic stability similar to Watson-Crick base pairs. This stability ensures the integrity of RNA secondary structures and promotes reliable translation of the genetic code.
- Essential for RNA secondary structure: Wobble base pairs play a fundamental role in the formation of RNA secondary structures. They contribute to the stability and folding of RNA molecules, enabling the proper functioning of RNA in various cellular processes.
- Faster dissociation and protein synthesis: The wobble base pairing allows faster dissociation of tRNA from mRNA during the translation process. This rapid dissociation promotes efficient protein synthesis by facilitating the movement of ribosomes along the mRNA template.
- Minimizing errors in genetic code interpretation: The existence of wobble minimizes the impact of certain errors in the genetic code. If a codon is misread during transcription, wobble allows the tRNA to still recognize and correctly translate the codon, maintaining the appropriate amino acid sequence during protein synthesis. This reduces the potential damage that can arise from occasional errors in the reading of the genetic code.
Quiz on Wobble Hypothesis
What does the Wobble Hypothesis explain?
a) The structure of DNA
b) The replication of DNA
c) The flexibility in the pairing of the third base of the codon
d) The synthesis of proteins
Who proposed the Wobble Hypothesis?
a) Rosalind Franklin
b) James Watson
c) Francis Crick
d) Maurice Wilkins
According to the Wobble Hypothesis, which position of the codon shows flexibility in base pairing?
a) First
b) Second
c) Third
d) Fourth
Which base can pair with multiple bases according to the Wobble Hypothesis?
a) Adenine
b) Cytosine
c) Guanine
d) Thymine
The Wobble Hypothesis helps to explain why:
a) There are more codons than amino acids
b) There are more amino acids than codons
c) Codons are always of fixed length
d) DNA is double-stranded
Which of the following is NOT a valid wobble pairing?
a) G-U
b) A-U
c) I-A
d) C-G
The Wobble Hypothesis reduces the need for:
a) Multiple DNA strands
b) Multiple types of amino acids
c) Multiple types of tRNAs
d) Multiple types of ribosomes
Inosine (I) can pair with which of the following bases?
a) Adenine
b) Cytosine
c) Both Adenine and Cytosine
d) Neither Adenine nor Cytosine
The Wobble Hypothesis is primarily associated with:
a) DNA replication
b) Transcription
c) Translation
d) DNA repair
The flexibility in base pairing, as proposed by the Wobble Hypothesis, occurs between:
a) mRNA codon and DNA template
b) mRNA codon and tRNA anticodon
c) tRNA anticodon and DNA template
d) tRNA anticodon and ribosomal RNA
FAQ
What is the wobble hypothesis?
The wobble hypothesis proposes that the base at the 5′ end of the anticodon in tRNA is not as strictly paired with the corresponding base in the mRNA codon, allowing for non-standard or wobble base pairings.
Why is it called the “wobble” hypothesis?
It is named the “wobble” hypothesis because the base at the wobble position is not spatially confined like the other two bases in the anticodon, allowing it to wobble or move unsteadily and form non-standard base pairs.
How does the wobble hypothesis explain degeneracy in the genetic code?
The wobble hypothesis suggests that the relaxed base-pairing rules at the third position of the codon allow a single tRNA molecule to recognize more than one codon. This accounts for the degeneracy or redundancy of the genetic code.
What are the main wobble base pairs?
The main wobble base pairs are guanine-uracil (G-U), hypoxanthine-uracil (I-U), hypoxanthine-adenine (I-A), and hypoxanthine-cytosine (I-C).
Why is hypoxanthine used in wobble base pairs?
Hypoxanthine is used to represent wobble base pairs because it is the nucleobase of inosine, which displays the true qualities of wobble by allowing for pairing with multiple bases in the original codon.
How does the wobble hypothesis impact protein synthesis?
The wobble base pairing allows for faster dissociation of tRNA from mRNA during protein synthesis, facilitating the movement of ribosomes and enhancing the efficiency of translation.
What role do wobble base pairs play in RNA secondary structure?
Wobble base pairs are fundamental in RNA secondary structure. They contribute to the stability and folding of RNA molecules, influencing their overall structure and function.
Does the wobble hypothesis affect the accuracy of the genetic code?
Yes, the wobble hypothesis helps to minimize errors in the interpretation of the genetic code. It ensures that even if there is a mismatch at the wobble position, the correct amino acid can still be incorporated during protein synthesis.
How does the wobble hypothesis impact the number of tRNA molecules needed?
The wobble hypothesis allows a single tRNA molecule to recognize multiple codons due to non-standard pairings. This broad specificity reduces the number of unique tRNA molecules required for translation.
How has the wobble hypothesis contributed to our understanding of molecular biology?
The wobble hypothesis has provided insights into the efficiency and accuracy of protein synthesis, RNA structure, and the functioning of the genetic code. It has enhanced our understanding of how cells optimize resources and maintain fidelity in the complex process of translating genetic information into functional proteins.
References
- https://microbenotes.com/the-wobble-hypothesis/
- https://en.wikipedia.org/wiki/Wobble_base_pair
- https://teaching.ncl.ac.uk/bms/wiki/index.php/Wobble_Hypothesis
- https://link.springer.com/referenceworkentry/10.1007%2F978-3-642-11274-4_1692
- https://www.biologydiscussion.com/genetics/wobble-hypothesis-with-diagram-genetics/65163
