Table of Contents
Principle of Transfection
According to the definition of transfection, the external genetic material must enter the cell via the cell membrane. It is essential to highlight that the genetic material, whether DNA or RNA, is negatively charged as a result of the proteins that surround it. Therefore, in an unmanipulated environment, exogenous DNA cannot cross the cell membrane. However, during transfection, the genetic material becomes coupled with positively charged molecules or is inserted straight into the nucleus of the host cell. This results in the elimination of the barrier for traversing the charged membrane. Chemical transfection and physical transfection can be distinguished according to the method employed to transmit exogenous genetic material, namely conjugation with cationic chemicals or insertion into the nucleus. As an example of this category, transduction falls under biological transfection, in which transfection is mediated by viral vectors.
Transfection Method
Here, we present a standard protocol for the transfection of adherent U2OS osteosarcoma cells with GFP plasmid DNA using a chemical transfection method in order to achieve transient expression of GFP in the host cell. This protocol has been successfully applied to a wide variety of adherent cell types. Depending on cell type, culture format, and transfection reagents used, the standard form of this protocol can be altered and optimised for a range of cell lines, taking into account the unique requirements of the method employed and the recommendations of the reagent manufacturer. For instance, when using an electroporation method with nonadherent cells, the number of cells and amount of transfected material must be significantly increased for a successful outcome, and if transfection efficiency is of utmost importance, co-transfection with a fluorescent marker that can be evaluated via immunofluorescence or flow cytometry may be considered. Fig. 1 depicts a schematic of the transfection workflow.
Materials required for Transfection
Nucleic acid preparation
- Use DNA and RNA of high quality. For this example, we will use commercially available enhanced green fluorescent protein (EGFP) plasmid DNA (e.g., pEGFP-N1 from Clontech). Nucleic acid should be of high quality; we use Qiagen plasmid purification kits as an example (Qiagen).
- Dilute plasmid with nuclease-free water or an appropriate buffer (e.g., 10mM Tris-HCl pH 8.5 (EB supplied with Qiagen kits)) to a standard concentration (e.g., 200ng/µL).
Cell culture and transfection reagents
- Unless otherwise specified, cells should be cultured at 37°C with 5% CO2 in a humidified incubator. Use cells with <50 passages and only quickly growing cells for transfection (unless unavoidable and thus use a transfection method that can be used for non-proliferating cells, e.g., retrovirus). Maintain uniform conditions for cell development and density for repeatability and standardisation objectives.
- Medium for cell culture supplemented with foetal calf serum (FCS) and antibiotic (if used), such as Dulbecco’s Modified Eagle Medium (DMEM) with 5% FCS and 1% Pen/Strep.
- On the day of or the day before transfection, seed cells in 6cm dishes (volume of complete growth medium 2mL). Depending on the cell culture format, the majority of manufacturers will include instructions on the recommended number of cells, amount of nucleic acid to be used for transfection, and ratio of nucleic acid to transfection reagent.
- Serum-free medium (e.g., Opti-MEM, Invitrogen).
- Transfection substance (e.g., GeneJuice, Novagen).
Western blotting
- Determine the protein concentration of the cell lysate using a typical Bradford test, for example. Protein loading buffer (for instance, 4x Laemmli sodium dodecyl sulphate (SDS) sample buffer with 2-beta-mercaptoethanol).
- Stacking and resolving SDS-PAGE (polyacrylamide gel electrophoresis) gels (in this instance, a 12% resolving gel was utilised, as the anticipated molecular weight of EGFP is 27kDa). In general, the percentage of acrylamide in a protein depends on its molecular weight.
- Note that the pH of the transfer buffer can be altered based on the isoelectric point (pI) of the protein of interest. As needed, we typically utilise Towbin transfer buffer pH 8.3 with 10% methanol and 0.1% SDS.
- Protein rung/molecular weight indicator (e.g., Protein Color Standard, New England Biolabs Inc).
- Gel apparatus and transfer equipment.
- Nitrocellose or polyvinylidene difluoride (PVDF) membrane (Immobilon, Millipore, UK).
- Ponceau S (Sigma-Aldrich).
- Blocking agent (e.g., 5% skimmed milk).
- Primary and secondary antibodies that are optimal.
Immunofluorescence
- Glass microscope slides with 13mm diameter coverslips (VWR International)
- Six-well dishes (Costar).
- 3.7% formaldehyde/phosphate-buffered saline (PBS) for fixing.
- 0.5% TritionX-100 in PBS to permeabilize cells.
- 5% FCS-PBS for blocking (if required).
- PBS/0.025% Tween is used to clean coverslips.
- Primary and secondary antibodies against the target protein.
- Example of a mounting medium: vectashield with 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories).
- Clear nail polish is used to seal coverslips.
- Box of opaque, humidified plastic for incubation procedures.
Transfection Protocols
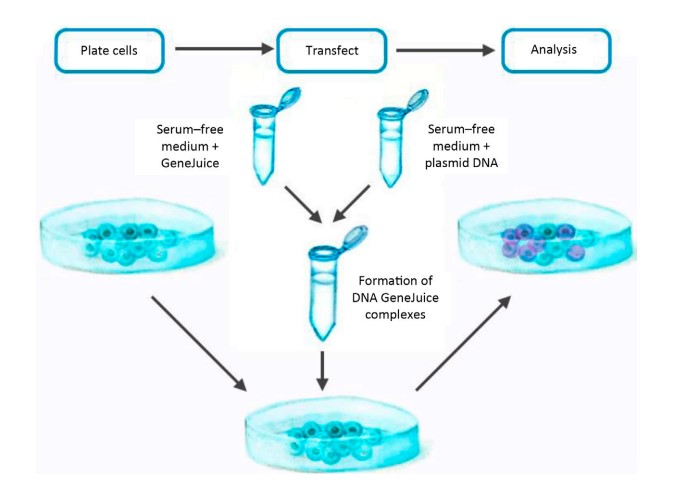
1. Transfection
a. Cell density
In general, a cell confluence between 40 and 60% is ideal. This varies according on individual choice, required culture time, and success with an optimised approach; a confluence as low as 20% can also result in successful transfection.
b. Ratio of transfection reagent to DNA
The majority of manufacturers will indicate this in their protocol. In this example, the transfection reagent is GeneJuice, which in our laboratory is employed at a concentration of 1.5µL per 1g of DNA plasmid. Note: Depending on the transfection material and conditions, the ratio of DNA to transfection reagent can range between 1:1 and 3:1. Depending on the technique, siRNA transfection reagents can be employed in ratios ranging from 2:1 to 4:1.
c. Transfection procedure
- Plate 10–20 × 105 U2OS cells per 6cm dish the day prior to transfection, and incubate at 37°C (5% CO2) overnight. Achieve between 20% and 40% cell confluence in order to conduct the experiment. Note: Include suitable controls beside your experiment. For instance, a control plasmid (e.g., an empty vector) or nontargeting siRNA should be selected to evaluate nonspecific effects of the test plasmid/siRNA under identical transfection conditions. In addition, a mock transfection (transfection reagent mix without nucleic acid) can be utilised as a control.
- For each plate to be transfected, pipette 500µL serum-free media into a sterile Eppendorf tube and add 1.5µL GeneJuice for each 1µg of DNA to be utilised. Vortex, then incubate at room temperature for 5 minutes. Note: We recommend preparing a master mix for all individual transfections by combining serum-free medium and the required amount of GeneJuice in a single tube before adding DNA to individual tubes (see below). This can reduce transfection efficiency variation.
- For each plate to be transfected, add the necessary amount of DNA to a fresh Eppendorf tube (e.g., 500ng and 1µg of pEGFP-N1 in Figures 11.2 and 11.3), followed by the serum-free medium/transfection reagent mixture, and mix gently by pipetting up and down three times (do not vortex). Incubate at ambient temperature for around 15 minutes (we generally find 5–15 minutes optimal).
- Drop by drop, add the transfection mixture to the separate dishes. Plates may be gently stirred, but they should not be whirled.
- If available, check plates daily using fluorescence microscopy. After a few hours, EGFP should become visible in transfected cells. Transfection efficiency can be estimated by measuring the ratio of fluorescent cells to nonfluorescing cells (compare the number of cells per field observed with conventional light microscopy to the number of cells per field observed with fluorescence to visualise EGFP). Other approaches for assessing transfection efficiency include fluorescence-activated cell sorting (FACS) and the addition of a marker of transfection efficiency whose expression levels can be evaluated, such as luciferase or -galactosidase.
- Cells can be extracted 48–72 hours after culturing. Cell pellets are resuspended in the proper lysis solution (we typically use TNN buffer (50mM Tris pH = 7.4, 5mM EDTA, 0.5% NP40, 150mM NACl with protease and phosphatase inhibitors)). Lysates must rest on ice for 20–30 minutes before to clarifying by centrifugation at 15,700 g for 10 minutes at 4°C. Lysates must be kept at 80°C until EGFP protein expression can be analysed by Western blotting.
2. Western blotting
a. SDS-PAGE gel electrophoresis and transfer
- Samples held at 80°C must be thawed, and the protein concentration of each sample must be measured using a standard protein quantitation assay (such as the Bradford reagent from Sigma-Aldrich).
- Determine the amount of protein you wish to load onto the gel (typically between 30 and 60 µg, depending on the protein) and add the necessary amount of lysate to a fresh tube before adding SDS sample buffer. Heat samples at 95°C for 5 minutes in a heat block, and remove any condensation by centrifuging quickly (e.g., 10 seconds) at room temperature. Now, your sample is prepared to be loaded onto the gel.
- Load samples in the selected running order and include a protein ladder in one gel lane to compare the relative molecular mass.
- Run the gels at 200V for 45 minutes, or until the loading dye has reached the gel’s bottom.
- Transfer gels into a transfer tank (per manufacturer’s instructions) so that proteins can be transferred electrophoretically onto a membrane (e.g., PVDF or nitrocellulose). At 4°C, transfers can operate at 400mA for 60–90 minutes, or at 150mA overnight.
- Remove membranes and stain them with Ponceau S to detect protein quickly. Wash membranes with distilled water so that protein bands can be seen clearly. Ponceau S is a reversible protein stain that can be quickly and readily removed prior to immunodetection via a fast PBS wash.
- Block membranes in 5% skim milk (w/v) in PBS containing 0.1% Tween-20 (SM-PBST) for 20 minutes (note that the choice of blocking can also vary; for instance, if you were interested in detecting a phospho-protein, bovine serum albumin (BSA) would be the preferred blocking agent; in addition, the use of Tris-buffered saline (TBS) instead of PBS throughout is recommended).
b. Incubation with primary antibody
- Add the primary antibody of choice to the needed volume of 5% SM-PBST and incubate overnight at 4°C. Primary antibody concentrations are typically between 1:500 and 1:1000, but can be adjusted during protocol optimization. Typically, the manufacturer’s datasheet is an adequate guidance.
- Cleanse membranes with PBST (e.g., a short wash of 1 minute should suffice).
c. Incubation with secondary antibody
- In 5% SM-PBST, add the appropriate secondary antibody. Typically, this is added at a 1:10,000 concentration.
- Incubate for 30–40 minutes at room temperature.
- Rinse membranes in PBST (e.g., three washes for 5 minutes each, but exact conditions can depend on the individual antibody and may be altered to accommodate for differences in background, strength of signals, etc.).
d. Detection of antibody complexes
- Enhanced chemiluminescence (ECL) is utilised to identify antibody/antigen complexes. This method necessitates the use of a darkroom and film developer. In addition, the reagents can be bought commercially or manufactured at home.
3. Immunofluorescence
a. Culture cells onto coverslips in six-well plates
- Inoculate roughly 1 105 cells per well with cells. Using 500ng EGFP DNA and 250µL of serum-free media, transfect according to the preceding technique.
- After the necessary period of growth (e.g., 24–72 hours), rinse cells twice with PBS.
b. Add 1 mL of 3.7% formaldehyde-PBS to each well and fix cells at room temperature for 10–15 minutes.
c. Remove fix and wash three times with PBS. d. Permeabilize cells with 0.5% TritonX-100 in PBS at room temperature for 2–5 minutes. g. Block cells for up to 30 minutes with 5% FCS-PBS at room temperature. Note: This step is optional. Depending on the antibody employed, blocking conditions may be changed or eliminated entirely.
d. To coverslips, add the main antibody. Add 20µL of the appropriate concentration of the main antibody. Incubate in a humidified dark box at 4°C overnight, or at ambient temperature for 30–60 minutes. Similar to Western blotting, check the manufacturer’s instructions for the recommended dilution; if employing an antibody for the first time without a recommendation, start with a dilution of 1:200 and adjust the concentration based on the results of successive trials.
e. After washing the slides 2–4 times with PBS/0.025% Tween-20, add the secondary antibody of choice. 30 minutes of incubation in the dark at room temperature. Four to six washes with PBS/0.025% Tween-20. If not otherwise specified, use secondary antibodies at a concentration of 1:500.
f. Mount coverslips (cell side down) onto glass slides with DAPI-containing mounting media. Clear nail polish is used to seal the edges of the coverslips.
g. Visualize slides with proper filters on an immunofluorescence microscope.
h. Store slides in dark boxes at 4°C.
Application of Transfection
There are numerous uses of transfection, including the ones listed below:
- It is an analytical tool used to examine the expression of genes or proteins in distinct cell types.
- Occasionally, siRNA transfection is utilised to reduce the expression of specific proteins.
- It enables the creation of recombinant proteins on a huge scale.
- Gene therapy relies on the transfection technique.
- It is used to silence genes
- It is utilised to establish stable cell lines.
