Table of Contents
What is cDNA cloning?
- cDNA cloning is a specialized technique in molecular biology that focuses on the isolation and study of specific genes of interest. The process begins with the extraction of mRNA from cells at a particular developmental stage or under specific conditions. This mRNA serves as a template for the synthesis of complementary DNA, commonly referred to as cDNA.
- Therefore, cDNA is essentially a DNA version of the mRNA, and its name, “copy DNA,” reflects this relationship. Unlike the original genomic DNA, cDNA only represents the genes that are actively expressed in the cell at the time of mRNA extraction. This makes cDNA cloning particularly valuable for studying gene expression patterns.
- The conversion of mRNA to cDNA is facilitated by an enzyme called reverse transcriptase. This enzyme synthesizes a single-stranded DNA copy of the mRNA template. Following this, another enzyme, DNA polymerase, is employed to generate the second DNA strand, resulting in a double-stranded cDNA molecule.
- Besides its role in synthesizing the complementary strand, DNA polymerase ensures the accuracy and fidelity of the cDNA sequence. Once the double-stranded cDNA is formed, it is integrated into a suitable vector, such as a plasmid or lambda phage, for further study and analysis.
- Then, the cDNA clone can be used for various applications, including gene expression studies, protein production, and functional analysis of genes. The emphasis on the function is crucial, as the cDNA clone represents the functional part of the genome, excluding introns and other non-coding regions.
- In summary, cDNA cloning is a precise and technical process that allows researchers to study the function and expression of specific genes. By utilizing specific enzymes and maintaining a clear and concise methodology, scientists can obtain valuable insights into the genetic basis of various biological processes and phenomena.
Definition of Complementary DNA or cDNA
Complementary DNA (cDNA) is a form of DNA synthesized from a messenger RNA (mRNA) template through the action of the enzyme reverse transcriptase. It represents the coding sequence of genes and is commonly used in gene cloning and expression studies.
cDNA Cloning Definition
cDNA cloning is a method used to clone expressed genes. It involves the reverse transcription of mRNA molecules to produce cDNA, which is then amplified and inserted into a vector for further studies or expression in a host organism. The primary advantage of cDNA cloning over genomic DNA cloning is that cDNA represents only the expressed genes, excluding introns and non-coding regions.
This technique is particularly useful for studying gene expression, producing recombinant proteins, and understanding the function of specific genes.
Principle of cDNA cloning/cdna synthesis principle
- The principle of cDNA cloning revolves around the conversion of mRNA molecules, which carry genetic information from the DNA to the ribosomes, back into a DNA form. This reverse process is essential for studying specific genes, especially in organisms with large amounts of non-coding DNA.
- To begin with, mRNA molecules are isolated from cells. These mRNA molecules represent the actively transcribed genes in a cell at a specific time. Therefore, by studying the cDNA derived from mRNA, researchers can gain insights into which genes are active under certain conditions.
- Once the mRNA is isolated, the enzyme reverse transcriptase is employed. This enzyme has the unique ability to synthesize a complementary DNA strand using an RNA template. The result of this synthesis is a single-stranded DNA molecule that mirrors the sequence of the mRNA, hence the term “complementary DNA” or cDNA.
- Following this, another enzyme, DNA polymerase, is introduced. Its function is to create a second, complementary DNA strand, resulting in a double-stranded cDNA molecule. This molecule is then ready for insertion into a vector, typically a bacterial plasmid.
- Besides the direct synthesis of cDNA, the concept also extends to the creation of cDNA libraries. These libraries are collections of cDNA clones that represent the entirety of genes expressed in a particular tissue or under specific conditions. There are two primary types of gene libraries: cDNA libraries and genomic DNA libraries. While genomic DNA libraries contain fragments of the entire genome, cDNA libraries specifically represent the genes that were actively being transcribed at the time of mRNA extraction.
- In summary, the principle of cDNA cloning is a detailed and sequential process that involves the reverse transcription of mRNA to produce complementary DNA. This technique allows for the study and analysis of gene expression and function in various organisms and under different conditions. The emphasis on the functions of the enzymes and the resulting cDNA molecules underscores the importance of this method in molecular biology research.
Steps of cDNA cloning
The process involves several detailed and sequential steps, each contributing to the successful cloning of complementary DNA (cDNA) from an mRNA template. Here are the steps involved in cDNA cloning:
- Isolation of mRNA: The first step in cDNA cloning is the extraction of messenger RNA (mRNA) from cells. This mRNA carries the genetic information for protein synthesis and represents the genes that are actively being transcribed in the cell at a specific time.
- Synthesis of the First Strand of cDNA: Once the mRNA is isolated, the enzyme reverse transcriptase is introduced. This enzyme synthesizes a complementary DNA strand using the mRNA as a template. The resulting product is a single-stranded cDNA molecule that mirrors the sequence of the mRNA.
- Synthesis of the Second Strand of cDNA: Following the creation of the first cDNA strand, the enzyme DNA polymerase comes into play. Its role is to synthesize the second, complementary DNA strand, resulting in a double-stranded cDNA molecule. This step ensures the stability and integrity of the cDNA for subsequent cloning processes.
- Cloning of cDNA: With the double-stranded cDNA in hand, the next step is its insertion into a suitable vector, typically a bacterial plasmid. This vector will serve as a vehicle to introduce the cDNA into host cells, allowing for its replication and expression.
- Introduction to Host Cells: The vector containing the cDNA is then introduced into host cells, commonly bacteria, through a process called transformation. These host cells will take up the vector and, in doing so, replicate and express the cDNA.
- Clone Selection: After transformation, it’s essential to select the host cells that successfully took up the cDNA-containing vector. This is done using various selection methods, ensuring that only the desired clones are retained for further study.
cDNA Cloning Protocol
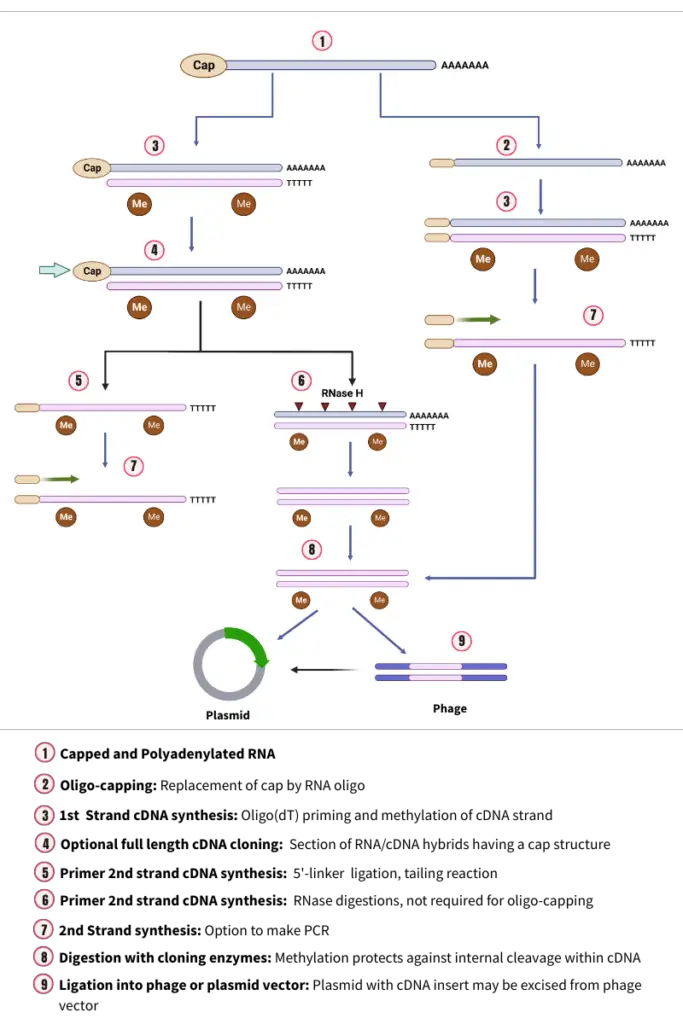
1. Isolation of mRNA
The process of cDNA cloning begins with the isolation of messenger RNA (mRNA) from the tissue containing the gene of interest. This step is crucial as mRNA carries the genetic information that will be used to create the complementary DNA (cDNA).
- Preparation of Crude Extract: Initially, a crude extract is prepared from the tissue that houses the desired gene. This extract serves as the primary source from which mRNA will be isolated.
- Purification: It’s essential that the extract be devoid of any contaminants such as proteins and polysaccharides. A pure mRNA sample ensures the accuracy and efficiency of subsequent steps in the cDNA cloning process.
- Oligo-dT Cellulose Chromatography: To further purify the mRNA, a technique called oligo-deoxythymine (oligo-dT) cellulose chromatography is employed. This method takes advantage of the unique structure of eukaryotic mRNAs, which possess a poly A tail (a sequence of adenine residues) at their 3′ end.
- Binding to Thymidine Residues: The poly A tail of the mRNA binds to a string of thymidine residues that are immobilized on cellulose. By controlling the conditions, the bound mRNA, also known as the poly (A)+ fraction, can be eluted or separated from the column.
- Enrichment of mRNA: To ensure a high concentration of mRNA, the poly (A)+ fraction is passed through the column multiple times. This repetitive process results in a fraction that is highly enriched for mRNA.
- Extraction of Specific mRNA: While the enriched fraction contains various mRNA sequences, specific techniques can be applied to isolate a particular mRNA species of interest.
- Confirmation of mRNA Sequence: After obtaining the desired mRNA fraction, it’s imperative to verify its content. This is achieved by translating the mRNA in vitro, or outside a living organism. The resulting polypeptides are then analyzed to confirm the presence of the sequence of interest.
2. Reverse transcriptases and first-strand cDNA synthesis
The synthesis of the first strand of complementary DNA (cDNA) is a pivotal step in the cDNA cloning process. This step involves the conversion of the previously isolated messenger RNA (mRNA) into a complementary DNA strand, setting the stage for the creation of a double-stranded cDNA molecule.
- Role of Reverse Transcriptase: Central to this process is an enzyme known as reverse transcriptase. This enzyme is unique as it is an RNA-dependent DNA polymerase. Its primary function is to transcribe the mRNA fraction, effectively copying it into the first strand of DNA.
- Primer Dependency: It’s essential to note that reverse transcriptase, like all DNA polymerases, requires a primer to initiate the synthesis. This primer provides the necessary 3′-OH group where the enzyme can add nucleotide residues.
- Use of Oligo-dT Primer: In the context of cDNA cloning, the most frequently employed primer is the oligo-dT. This primer, which consists of 12-18 nucleotides in length, is specifically designed to bind to the poly (A) tract located at the 3′ end of mRNA molecules. Therefore, the oligo-dT primer serves as a starting point for the reverse transcription process, ensuring the accurate replication of the mRNA sequence.
- Formation of RNA-DNA Hybrid: As the reverse transcriptase progresses, it synthesizes a complementary DNA strand, resulting in the formation of an RNA-DNA hybrid. This hybrid structure contains both the original mRNA strand and its newly synthesized DNA counterpart.
- Destruction of the RNA Strand: Before moving on to the synthesis of the second cDNA strand, it’s crucial to eliminate the RNA strand from the hybrid. This is achieved through a process called alkaline hydrolysis, which effectively breaks down the RNA component, leaving behind only the first strand of cDNA.
3. Synthesis of Second-strand cDNA synthesis
Following the synthesis of the first strand of cDNA, the next pivotal step is the creation of the second complementary DNA strand. This process ensures the formation of a complete double-stranded cDNA molecule, which is essential for subsequent cloning procedures. The synthesis of the second strand can be achieved through two primary techniques:
- Self-Priming cDNA Synthesis:
- Mechanism: In the self-priming method, the cDNA:mRNA hybrid obtained from the first strand synthesis undergoes denaturation. This denaturation facilitates the synthesis of the second strand on the single-stranded cDNA using the Klenow fragment of DNA polymerase I.
- Hairpin Structure: A transient hairpin structure forms at the 3′ end of the single-stranded DNA. This structure serves as a primer, enabling the Klenow fragment of Escherichia coli DNA polymerase I to initiate the synthesis of the second cDNA strand.
- S1 Nuclease Digestion: To ensure the proper formation of the double-stranded cDNA, the hairpin loop and any single-stranded overhangs at the other end are digested using the single-strand-specific S1 nuclease.
- Outcome: The final product of this method is a collection of double-stranded, blunt-ended DNA molecules that are complementary to the original mRNA fraction.
- Replacement Synthesis:
- Template Role: In the replacement synthesis method, the cDNA:mRNA hybrid serves as a template for a nick translation reaction.
- Creation of Nicks and Gaps: RNase H introduces nicks and gaps in the mRNA strand of the hybrid, leading to the formation of a series of RNA primers.
- Role of E. coli DNA Polymerase I: These RNA primers are utilized by E. coli DNA polymerase I to synthesize the second strand of cDNA.
- Advantages: This technique offers several benefits. It is highly efficient and can be directly executed using the products of the first strand reaction. Moreover, it eliminates the requirement for nuclease S1 to cleave the single-stranded hairpin loop in the double-stranded cDNA, streamlining the process.
4. Cloning of cDNA
Cloning of cDNA is a pivotal step in the process of cDNA synthesis, aiming to integrate the synthesized cDNA into a suitable vector for further analysis and applications. This step ensures that the cDNA is stably maintained and can be propagated in a host organism, typically E. coli. The procedure for cloning cDNA can be elucidated as follows:
- Homopolymeric Tract Addition:
- Overview: The most prevalent method for cloning cDNAs involves the strategic addition of complementary homopolymeric tracts to both the double-stranded cDNA and the chosen plasmid vector.
- cDNA Tailing: Using the enzyme terminal transferase, strings of cytosine residues are appended to the cDNA, resulting in the formation of oligo-dC tails on the 3′ ends of the cDNA molecule.
- Vector Preparation:
- Plasmid Modification: Concurrently, a plasmid vector is prepared by cleaving it at a unique restriction endonuclease site. Subsequently, this plasmid is tailed with oligo-dG, creating complementary sequences to the tailed cDNA.
- Hybrid Molecule Formation:
- Hydrogen Bonding: The tailed cDNA and the modified plasmid vector are then combined, allowing for hydrogen bonding between the complementary homopolymers (oligo-dC tails of cDNA and oligo-dG tails of the plasmid).
- Resultant Structure: This interaction culminates in the creation of open circular hybrid molecules. These molecules possess the unique capability of transforming E. coli cells.
In essence, the cloning of cDNA is a meticulously orchestrated process that ensures the stable integration of cDNA into a suitable vector. This integration facilitates the propagation and study of the cDNA in a host organism, providing valuable insights into the gene of interest.
5. Introduction to host cells
The process of introducing the recombinant DNA into host cells, often E. coli, is a critical step in the molecular cloning workflow. This step ensures that the cloned DNA is propagated and expressed within a living organism, allowing for further analysis and applications. Here’s a detailed breakdown of this step:
- Utilizing Recombinant Plasmids:
- Choice of Host: The E. coli K-12 strain is predominantly chosen as the host organism for transformation.
- Role of Plasmids: Recombinant plasmids serve as the vehicle to introduce foreign DNA into the host cells.
- Uptake of Plasmid Molecules:
- Treatment with Calcium Chloride: E. coli cells are treated with calcium chloride, a process that makes them competent to take up plasmid molecules from their surrounding environment.
- Gap Repair: Following the uptake, any gaps present in the recombinant plasmid are efficiently repaired by the host cells.
- Isolation of Transformed Bacteria:
- Antibiotic Resistance as a Marker: The transformed bacteria can be distinguished from the non-transformed ones based on their resistance to specific antibiotics.
- Multiple Resistance Genes: Most cloning plasmids are equipped with two antibiotic resistance genes. However, one of these genes gets disrupted during the cloning process. Therefore, the intact gene serves as a marker for successful transformation.
- Example with pBR322: In the context of the pBR322 plasmid, cloning into the unique PstI site disrupts the ampicillin resistance gene, leaving the tetracycline resistance gene unaffected. As a result, bacteria that have been successfully transformed with a recombinant plasmid will exhibit sensitivity to ampicillin but will remain resistant to tetracycline.
In summary, the introduction of recombinant DNA into host cells is a meticulously executed process that ensures the successful propagation of cloned DNA within a living organism. This step is pivotal for subsequent analyses and applications related to the gene of interest.
6. Clone selection
Clone selection is a pivotal step in the molecular cloning process. After the introduction of recombinant DNA into host cells, it becomes imperative to identify and select the specific clones that carry the desired DNA sequence. Here’s an in-depth look at this crucial step:
- Initial Antibiotic Resistance Selection:
- Distinguishing Clones: While the antibiotic resistance selection can determine which clones possess a recombinant plasmid, it doesn’t specify the nature of the insert. Given that the cloning process often starts with a diverse population of mRNA sequences, there could be thousands of different inserts present.
- Challenges in Selection:
- Variety of Inserts: The primary challenge lies in selecting clones that carry the specific sequence of interest from the myriad of possibilities.
- Selection Based on Protein Expression:
- Expression as a Marker: If the gene of interest is expressed in the host cells, it offers a straightforward selection criterion.
- Screening Methods: The presence of the desired protein can be detected in multiple ways:
- Phenotypic Observation: The protein might confer a specific phenotype to the bacteria, making it distinguishable.
- Protein Detection Techniques: Various methods, often rooted in immunological or enzymological techniques, can be employed to detect the protein.
- Alternative Selection Methods:
- In Cases of Non-expression: If the desired gene isn’t expressed as a protein, alternative strategies must be employed.
- Nucleic Acid Hybridization: This method is particularly useful for identifying specific DNA sequences within a mixture. It involves the use of a labeled complementary DNA or RNA strand to pinpoint the location of a specific DNA sequence in a sample.
- Further Identification:
- Post Genomic DNA Cloning: Once the desired clones are selected, the identification of the specific gene or sequence can be further discussed, especially in the context of genomic DNA cloning.
Why cDNA is used for cloning?
- Representation of Expressed Genes: cDNA is synthesized from mRNA, which means it represents genes that are actively being expressed in the cell at the time of mRNA extraction. This allows researchers to specifically study genes that are functional and being translated into proteins.
- Absence of Introns: Unlike genomic DNA, cDNA does not contain introns because it is derived from mRNA, which has already undergone splicing. This makes cDNA more suitable for expression in prokaryotic systems like bacteria, which do not have the machinery to splice out introns.
- Simpler Sequence: Since cDNA represents only the coding sequences of genes (exons), it is often shorter and simpler to work with than genomic DNA.
- Expression Studies: cDNA libraries can be used to study gene expression patterns in different tissues, developmental stages, or under different conditions. By comparing the cDNA libraries from different sources, researchers can identify which genes are upregulated or downregulated under specific conditions.
- Recombinant Protein Production: cDNA can be cloned into expression vectors and introduced into host organisms like bacteria, yeast, or mammalian cells to produce the protein encoded by the cDNA. This is particularly useful for producing large quantities of a specific protein for research, therapeutic, or industrial purposes.
- Functional Genomics: cDNA cloning allows researchers to study the function of specific genes. By expressing the cDNA in a model organism, researchers can observe the phenotype or function associated with that gene.
- Comparative Genomics: cDNA sequences can be used to compare genes across different species, providing insights into evolutionary relationships and the conservation of gene function.
- Probe for Genomic Libraries: cDNA can be labeled and used as a probe to identify corresponding genomic sequences in genomic DNA libraries.
Application of cDNA
- Gene Expression Analysis: One of the primary uses of cDNA is to analyze gene expression patterns. For instance, cDNA microarrays can be employed to investigate gene expression patterns in specific tissues or under certain conditions. By comparing the expression levels of genes in different samples, researchers can identify which genes are upregulated or downregulated under specific circumstances.
- EST Sequencing: Expressed Sequence Tags (ESTs) are short sequences derived from cDNA. These ESTs provide a snapshot of gene expression in a particular tissue at a specific time. By sequencing ESTs, researchers can gain insights into the genes that are active in a given sample.
- cDNA Libraries: cDNA libraries represent a collection of cDNAs from a particular tissue or under a specific condition. These libraries are invaluable resources for studying gene expression and function. They allow researchers to identify and isolate genes based on their expression patterns.
- Protein Production: cDNA can be used to produce proteins in a laboratory setting. By inserting cDNA into an expression vector and introducing it into a host organism, researchers can produce large quantities of a specific protein. This is particularly useful for studying protein function or producing therapeutic proteins.
- Functional Genomics: cDNA plays a pivotal role in functional genomics, where the function of individual genes is studied. By introducing cDNA into cells, researchers can observe the effect of overexpressing a particular gene, providing insights into its function.
- Comparative Genomics: cDNA sequences can be compared across different species to study evolutionary relationships and identify conserved genetic elements.
- Mutation Analysis: cDNA can be used to study mutations in genes. By comparing the cDNA sequence of a gene from a healthy individual with that from an affected individual, researchers can identify mutations that may be responsible for diseases.
Application of cDNA Cloning
cDNA cloning is a pivotal technique in molecular biology that allows for the isolation and amplification of specific genes of interest. The application of cDNA cloning has numerous implications and benefits in scientific research and biotechnology:
- Expression of Proteins: cDNA cloning enables the expression of proteins in host organisms. By introducing the cDNA of a specific gene into an expression vector and then into a host organism, the protein encoded by that gene can be produced in large quantities. This is particularly useful for producing therapeutic proteins or for studying the function of specific proteins.
- Gene Function Analysis: Through cDNA cloning, researchers can study the function of specific genes. By introducing the cDNA clone into cells and observing the resultant phenotype, insights into the role of the gene in cellular processes can be gleaned.
- Comparative Genomics: cDNA clones allow for the comparison of genes across different species. This can provide insights into evolutionary processes and the conservation of gene function across species.
- Therapeutic Applications: cDNA clones can be used in gene therapy, where a functional gene is introduced into cells to replace or supplement a non-functional or absent gene. This has potential therapeutic applications for genetic disorders.
- Molecular Markers: cDNA clones can serve as molecular markers for mapping and sequencing genomes. They can help in identifying genes associated with specific traits or diseases.
- Transgenic Organisms: cDNA clones are instrumental in the creation of transgenic organisms. By introducing a specific cDNA clone into an organism, researchers can produce organisms with desired traits or characteristics.
- Disease Research: cDNA clones can be used to study the genes associated with specific diseases. By understanding the genes and their functions, researchers can develop targeted treatments and therapies.
FAQ
What is cDNA cloning?
cDNA cloning is a molecular biology technique used to isolate and amplify specific genes from an organism’s mRNA to produce complementary DNA (cDNA) and then insert it into a cloning vector for further study or expression.
How is cDNA different from genomic DNA?
cDNA is synthesized from mRNA and represents only the expressed genes of an organism, excluding introns. Genomic DNA, on the other hand, contains both coding (exons) and non-coding (introns) regions of genes.
Why is cDNA preferred for cloning over genomic DNA?
cDNA is derived from mRNA and lacks introns, making it suitable for expression in prokaryotic systems like bacteria that cannot process introns. It also represents only actively expressed genes, simplifying the study of gene expression.
How is cDNA synthesized?
cDNA is synthesized using the enzyme reverse transcriptase, which converts mRNA into its DNA complement. This process often uses an oligo-dT primer that binds to the poly-A tail of mRNA.
What is a cDNA library?
A cDNA library is a collection of cDNA clones representing the mRNA transcripts of a particular tissue, developmental stage, or condition. It provides a snapshot of gene expression at a specific time and place.
Why are cDNA libraries important?
cDNA libraries allow researchers to study gene expression patterns, identify novel genes, and produce recombinant proteins. They are essential tools for functional genomics and proteomics studies.
Can cDNA be used to express proteins in bacteria?
Yes, since cDNA lacks introns and represents the coding sequence of genes, it can be cloned into bacterial expression vectors and used to produce proteins in bacterial systems.
What are the applications of cDNA cloning?
cDNA cloning has various applications, including studying gene expression, producing recombinant proteins, functional genomics research, comparative genomics, and therapeutic drug development.
How is a cDNA clone different from a genomic DNA clone?
A cDNA clone contains only the exonic (coding) regions of a gene, while a genomic DNA clone includes both exonic and intronic (non-coding) regions.
What challenges might one encounter in cDNA cloning?
Challenges in cDNA cloning can include inefficient reverse transcription, incomplete cDNA synthesis, contamination with genomic DNA, and difficulty in cloning large cDNA fragments.
