Table of Contents
What is Bacterial Secretion Systems?
- Bacterial secretion systems are intricate protein assemblies embedded in bacterial cell membranes, responsible for the transport of substances, predominantly proteins. These systems are pivotal for pathogenic bacteria, enabling them to secrete virulence factors essential for invading host cells.
- The secretion processes can be broadly categorized into two mechanisms. The first is a direct, one-step mechanism where proteins are transported from the bacterial cytoplasm straight through the cell membrane into the host cell. The second is a more complex two-step process where proteins are first moved to the periplasmic space before being secreted into the host cell.
- The classification of these secretion systems is multifaceted, with distinctions often made between Gram-negative diderm bacteria and Gram-positive monoderm bacteria. However, this classification is intricate, with at least eight types specific to Gram-negative bacteria, four to Gram-positive bacteria, and two shared by both. Further complexities arise when considering diderm bacteria with lipopolysaccharide versus those with mycolic acid.
- A fundamental function in prokaryotic cells is the transport of proteins to various cellular compartments, the surrounding environment, or into other cells, a phenomenon termed protein secretion. Prokaryotes have evolved diverse mechanisms for protein transport, largely facilitated by specialized protein secretion systems.
- These systems are indispensable for bacterial growth, playing roles in a myriad of cellular processes. While some secretion systems are ubiquitous, secreting a plethora of substrates, others are more specific, dedicated to secreting a limited set of proteins.
- In certain scenarios, these systems enable bacterial pathogens to manipulate their host to create a conducive environment for replication. At other times, they help bacteria adapt to specific environmental niches, often by secreting proteins that provide a competitive edge against other microorganisms.
- The design of bacterial secretion systems can vary based on the number of membranes their protein substrates must traverse. Some systems facilitate protein transport across a single membrane, while others can transport proteins across two or even three membranes.
- Given the specificity of some of these systems in bacterial pathogens, they present potential targets for the development of novel antimicrobials, enhancing our existing arsenal of antibiotics.
- Several bacterial secretion systems, including the Type III to Type VII Secretion Systems, have been extensively studied. This discourse offers a concise overview of these systems, emphasizing their canonical features and structural-functional relationships.
- It is noteworthy that the host’s innate immune system can detect and respond to these protein secretion systems during infection, underscoring their significance in host-pathogen interactions.
- In conclusion, bacterial secretion systems are sophisticated molecular machines that play crucial roles in bacterial physiology and pathogenesis. Their intricate designs and functions make them subjects of intense scientific research, with potential implications for therapeutic interventions.
Definition of Bacterial Secretion Systems
Bacterial secretion systems are specialized protein complexes in bacterial cell membranes that transport substances, primarily proteins, from the bacterial cell to its environment or into host cells, facilitating processes such as pathogenesis and environmental adaptation.
Classes of bacterial protein secretion systems
Bacterial protein secretion systems are intricate molecular assemblies that facilitate the transport of proteins across bacterial membranes. These systems are classified based on various criteria, including the secretion signal, the number of steps involved in secretion, the state of the substrate (folded or unfolded), the number of membranes crossed, and their presence in Gram-positive or Gram-negative bacteria. Here, we delineate the primary classes of bacterial protein secretion systems:
- Sec System:
- Secretion Signal: N-terminus
- Steps in Secretion: 1
- Folded Substrates: No
- Number of Membranes: 1
- Bacterial Type: Both Gram-positive and Gram-negative
- Tat System:
- Secretion Signal: N-terminus
- Steps in Secretion: 1
- Folded Substrates: Yes
- Number of Membranes: 1
- Bacterial Type: Both Gram-positive and Gram-negative
- Type I Secretion System (T1SS):
- Secretion Signal: C-terminus
- Steps in Secretion: 1
- Folded Substrates: No
- Number of Membranes: 2
- Bacterial Type: Gram-negative
- Type II Secretion System (T2SS):
- Secretion Signal: N-terminus
- Steps in Secretion: 2
- Folded Substrates: Yes
- Number of Membranes: 1
- Bacterial Type: Gram-negative
- Type III Secretion System (T3SS):
- Secretion Signal: N-terminus
- Steps in Secretion: 1-2
- Folded Substrates: No
- Number of Membranes: 2-3
- Bacterial Type: Gram-negative
- Type IV Secretion System (T4SS):
- Secretion Signal: C-terminus
- Steps in Secretion: 1
- Folded Substrates: No
- Number of Membranes: 2-3
- Bacterial Type: Gram-negative
- Type V Secretion System (T5SS):
- Secretion Signal: N-terminus
- Steps in Secretion: 2
- Folded Substrates: No
- Number of Membranes: 1
- Bacterial Type: Gram-negative
- Type VI Secretion System (T6SS):
- Secretion Signal: No known secretion signal
- Steps in Secretion: 1
- Folded Substrates: Unknown
- Number of Membranes: 2-3
- Bacterial Type: Gram-negative
- SecA2 System:
- Secretion Signal: N-terminus
- Steps in Secretion: 1
- Folded Substrates: No
- Number of Membranes: 1
- Bacterial Type: Gram-positive
- Sortase System:
- Secretion Signal: N-terminus (Sec) and C-terminus (cws)
- Steps in Secretion: 2
- Folded Substrates: Yes
- Number of Membranes: 1
- Bacterial Type: Gram-positive
- Injectosome System:
- Secretion Signal: N-terminus
- Steps in Secretion: 2
- Folded Substrates: Yes
- Number of Membranes: 1
- Bacterial Type: Gram-positive
- Type VII Secretion System (T7SS):
- Secretion Signal: C-terminus
- Steps in Secretion: 1
- Folded Substrates: Yes
- Number of Membranes: 1-3
- Bacterial Type: Gram-positive
In summary, bacterial protein secretion systems are diverse and tailored to the specific needs of bacteria, playing pivotal roles in their physiology, pathogenesis, and interaction with their environment. Each system possesses unique characteristics that determine its function and specificity in protein transport.
Secretion Across The Cytoplasmic Membrane
Bacterial cells employ intricate mechanisms to transport proteins across their cytoplasmic membrane. This transport is pivotal for the cell’s interaction with its environment, pathogenicity, and overall physiology. Two primary pathways, the Sec and Tat pathways, are responsible for this translocation.
- The Sec Secretion Pathway:
- Mechanism: The Sec pathway predominantly translocates proteins in their unfolded state. It comprises three main components: a protein targeting component, a motor protein, and the SecYEG translocase, an integrated membrane channel. Some Gram-positive bacteria also produce Sec accessory proteins that aid in the secretion of specific proteins.
- Function: While the Sec pathway secretes a myriad of proteins, it is noteworthy for transporting virulence factors in certain bacterial pathogens, such as Vibrio cholerae, Klebsiella pneumoniae, and Yersinia enterocolitica.
- Export Mechanism: The Sec pathway utilizes a hydrophobic signal sequence located at the N-terminus of the protein. This sequence is recognized by the SecB protein in many Gram-negative bacteria. SecB acts as a chaperone, binding to pre-secretory proteins, preventing them from folding, and subsequently delivering them to SecA. SecA guides these proteins to the SecYEG channel and also functions as an ATPase, providing the energy required for protein translocation.
- The SRP Pathway:
- Mechanism: The SRP pathway, a subset of the Sec system, is responsible for transporting proteins that are destined to remain in the inner membrane. It employs a co-translational mechanism, coupling protein translation with secretion.
- Function: This pathway is essential for transmembrane proteins, which often contain hydrophobic domains that are unstable in the cytoplasm. The SRP particle, comprising a small 4.5S RNA bound to the Ffh protein, plays a pivotal role in this pathway.
- Export Mechanism: During secretion, the SRP first binds to the transmembrane domain of proteins as they emerge from the ribosome. It then binds to the docking protein FtsY, which delivers the ribosome-protein complex to the SecYEG channel. The protein’s translation then propels its secretion through the channel.
- The Tat Secretion Pathway:
- Mechanism: Unlike the Sec pathway, the Tat pathway primarily secretes proteins in their folded state. This is crucial as some proteins, especially those with post-translational modifications, need to be folded and modified in the cytoplasm before secretion.
- Function: The Tat pathway is essential for both pathogenic and non-pathogenic bacteria. For instance, certain pathogenic bacteria, including Pseudomonas aeruginosa and Yersinia pseudotuberculosis, rely on a functional Tat pathway for virulence.
- Export Mechanism: The Tat pathway comprises 2-3 subunits: TatA, TatB, and TatC. In Escherichia coli, TatB and TatC bind the signal peptide of Tat-secreted proteins and then recruit TatA, which forms the membrane-spanning channel. The Tat signal sequence contains a pair of “twin” arginines in the motif S-R-R at the N-terminus of the folded protein.
In conclusion, bacterial protein secretion across the cytoplasmic membrane is a complex process that is essential for the cell’s survival, interaction with its environment, and pathogenicity. The Sec and Tat pathways, with their distinct mechanisms and functions, play a central role in this intricate process.
The SecB Pathway in Bacterial Protein Secretion
The SecB pathway is a subset of the broader Sec secretion system, specifically tailored for the transport of certain proteins across the bacterial cytoplasmic membrane. Here’s a detailed look into the SecB pathway:
- Role and Function:
- Chaperone Activity: SecB acts as a chaperone protein. Its primary function is to bind to pre-secretory proteins, preventing them from prematurely folding in the cytoplasm. This ensures that these proteins remain in an unfolded state, which is essential for their subsequent translocation across the membrane.
- Mechanism:
- Recognition: Proteins destined for transport to the periplasm or outside of the cell via the SecB pathway contain a specific, removable signal sequence. This sequence is recognized by the SecB protein.
- Binding and Delivery: After binding to the target pre-secretory proteins, SecB delivers them to another protein called SecA.
- Interaction with SecA: SecA plays a dual role. It guides the proteins to the SecYEG channel, a membrane-integrated conducting channel. Additionally, SecA functions as an ATPase, providing the energy required for the translocation of proteins across the membrane.
- Signal Sequence Cleavage: Before the proteins are transported through the SecYEG channel, a protease cleaves off the SecB signal sequence. Once inside the periplasm, the proteins fold into their functional forms.
- Destination:
- Periplasmic Retention: Many proteins transported by the SecB system remain in the periplasm, where they perform various functions.
- Extracellular Transport: Some proteins, after being delivered to the periplasm by the SecB pathway, are transported outside the bacterial cell. This extracellular transport often requires the assistance of other secretion systems, such as the Type II and Type V secretion systems.
- Significance:
- The SecB pathway is particularly important in many Gram-negative bacteria. It ensures that proteins, especially those that need to be secreted into the periplasm or outside the cell, are correctly targeted and transported without premature folding.
In essence, the SecB pathway is a crucial component of the bacterial protein secretion machinery, ensuring that specific proteins are efficiently and correctly transported to their designated locations.
Secretion Mechanism through the Tat pathway
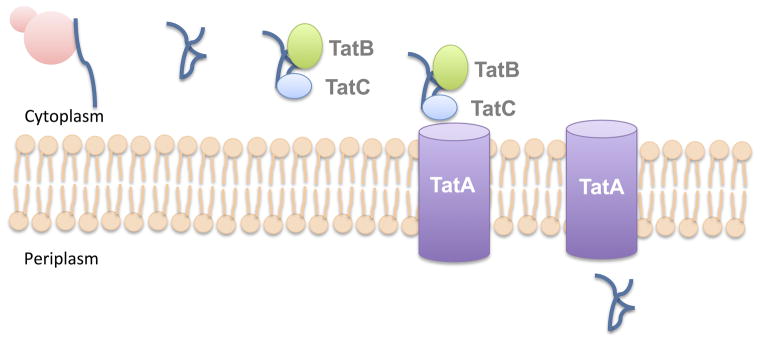
The Tat pathway is a specialized bacterial secretion mechanism responsible for the translocation of folded proteins across the cytoplasmic membrane. Unlike other secretion systems that transport unfolded proteins, the Tat pathway is unique in its ability to handle fully folded substrates. The intricacies of this pathway can be delineated as follows:
- Components of the Tat Pathway: The pathway is orchestrated by 2–3 integral components, namely TatA, TatB, and TatC. Their collective action ensures the efficient and accurate transport of proteins.
- Recognition of the Signal Peptide: In Gram-negative bacteria, the proteins destined for the Tat pathway are characterized by a specific N-terminal signal peptide. This peptide is unique due to the presence of a “twin” arginine motif. The proteins bearing this motif are recognized and bound by the TatB and TatC components.
- Formation of the Translocation Channel: Upon binding the folded secretion substrates, TatB and TatC engage TatA. This recruitment prompts TatA to localize to the cytoplasmic membrane, where it assembles to form a dedicated translocation channel.
- Protein Translocation: With the channel in place, the folded proteins are ushered across, making their way into the periplasmic space.
- Destination of Translocated Proteins in Gram-negative Bacteria: Once inside the periplasm of Gram-negative bacteria, the proteins have two potential fates. They can either reside within the periplasm or undergo further export out of the bacterial cell, facilitated by the Type II Secretion System (T2SS).
In summary, the Tat pathway is a remarkable bacterial secretion system, adept at transporting folded proteins. Its precise mechanisms and components underscore the complexity and adaptability of bacterial secretion processes, offering valuable insights into microbial physiology and potential therapeutic interventions.
Export Mechanism through the Sec pathway
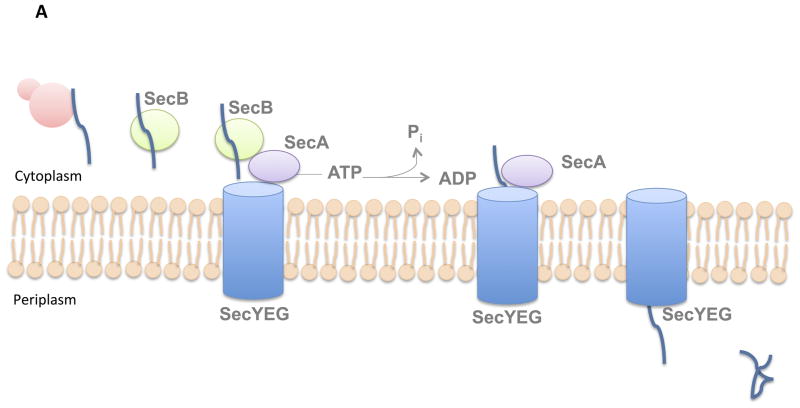
The Sec pathway is a fundamental protein export mechanism in bacteria, responsible for the translocation of unfolded proteins across the cytoplasmic membrane. Depending on their final destination, proteins transported via this pathway can either integrate into the inner membrane or be dispatched into the periplasm. For Gram-negative bacteria, an additional step may ensue, where periplasmic proteins are extracellularly released through another secretion system. The Sec pathway operates through two distinct mechanisms:
- Post-translational Translocation (Periplasmic/Extracellular Proteins):
- Signal Sequence Recognition: Proteins designated for the periplasm or extracellular release possess a removable signal sequence. This sequence is identified by the SecB protein.
- Role of SecB: SecB binds to these pre-secretory proteins, inhibiting their premature folding. Concurrently, it shuttles its substrates to SecA.
- Function of SecA: SecA performs dual roles. Firstly, it navigates proteins to the SecYEG channel. Secondly, it acts as an ATPase, supplying the energy required for protein translocation.
- Translocation and Folding: After traversing the SecYEG channel, proteins undergo folding within the periplasm.
- Co-translational Translocation (Inner Membrane Proteins):
- Signal Sequence and SRP: Proteins destined for the inner membrane have a signal sequence that attracts the Signal Recognition Particle (SRP).
- SRP and Ribosome Interaction: As proteins are being translated, SRP latches onto target proteins emerging from the ribosome. This interaction recruits the docking protein, FtsY.
- Delivery to SecYEG Channel: FtsY facilitates the transfer of the ribosome-protein complex to the SecYEG channel.
- Translocation and Integration: As the nascent protein is translocated across the channel, its transmembrane domain slips through the channel’s side, integrating into the membrane.
In essence, the Sec pathway is a sophisticated system that ensures precise protein localization, whether it’s within the membrane, the periplasm, or beyond. Understanding this pathway is crucial for insights into bacterial physiology and potential therapeutic strategies.
Protein Secretion By Gram-negative Bacteria
Gram-negative bacteria have evolved intricate systems to transport proteins across their complex cell envelopes, which consist of an inner and outer membrane separated by the periplasmic space. These protein secretion systems play pivotal roles in bacterial survival, adaptation, and pathogenesis. This article delves into the mechanisms of protein secretion in Gram-negative bacteria, emphasizing their structural and functional diversity.
- Overview of Protein Secretion Challenges: Gram-negative bacteria face the unique challenge of transporting proteins across two or, in certain instances, three phospholipid membranes to reach their extracellular destinations. Some proteins first reach the periplasm via the Sec or Tat secretion pathways and then cross the outer membrane using another transport system, a process termed Sec- or Tat-dependent protein secretion. Conversely, many proteins bypass the periplasmic step, being secreted directly through channels spanning both membranes, a phenomenon known as Sec- or Tat-independent secretion.
- The Type I Secretion System (T1SS): T1SSs, found in numerous Gram-negative bacteria, facilitate the one-step transport of their substrates across both bacterial membranes. Distinctively, T1SSs resemble ATP-binding cassette (ABC) transporters, which typically expel small molecules. These systems comprise three core components: an inner membrane ABC transporter, a membrane fusion protein (MFP), and an outer membrane factor (OMF). The ABC transporter, energized by ATP hydrolysis, recognizes and transports substrates, while the MFP bridges the inner and outer membranes. The OMF, often the versatile protein TolC, forms the outer membrane pore.
- The Type II Secretion System (T2SS): T2SSs, conserved across most Gram-negative bacteria, transport folded proteins from the periplasm to the extracellular milieu. These systems consist of multiple components, including an outer-membrane complex, an inner-membrane platform, a secretion ATPase, and a pseudopilus. The pseudopilus, resembling type IV pili, is believed to push substrates through the outer membrane channel in a piston-like manner.
- The Type III Secretion System (T3SS): T3SSs, present in many Gram-negative pathogens, are often described as “injectisomes” due to their needle-like structure. These systems can transport a plethora of effector proteins across bacterial membranes and directly into eukaryotic cells. The T3SS comprises a basal body, a needle component, and a translocon. The needle, with a hollow core, facilitates the passage of effectors, while the tip complex and translocon are crucial for sensing host cells and regulating secretion.
- The Type IV Secretion System (T4SS): T4SSs, evolutionarily linked to bacterial DNA conjugation systems, can transport diverse substrates, including proteins and DNA-protein complexes. These systems can transfer substrates to a variety of target cells, including other bacteria and eukaryotic cells. Despite their functional diversity, T4SSs share common components and operational principles.
- The Type V Secretion System: The Type V secretion system (T5SS) is unique among the bacterial secretion systems. It is often referred to as the autotransporter system due to its distinct mechanism of action. T5SSs are primarily found in Gram-negative bacteria and are responsible for the transport of large proteins across the outer membrane. These proteins, known as autotransporters, possess a characteristic domain structure: an N-terminal signal sequence, a passenger domain, and a C-terminal translocator domain. The process begins with the Sec-dependent transport of the autotransporter protein to the periplasm. Once in the periplasm, the translocator domain facilitates the formation of a β-barrel structure in the outer membrane. The passenger domain, often an enzyme or adhesin, is then translocated through this barrel to the cell surface. Some variations of the T5SS involve multiple proteins working in tandem to achieve secretion.
- The Type VI Secretion System: The Type VI secretion system (T6SS) is a complex, multi-component system found in many Gram-negative bacteria. It functions like a molecular crossbow, allowing bacteria to inject toxic effector proteins directly into neighboring bacterial or eukaryotic cells. Structurally, the T6SS resembles an inverted phage tail spike, with a contractile sheath surrounding an inner tube. Upon activation, the sheath contracts, propelling the inner tube and associated effector proteins out of the bacterial cell and into the target cell. This system plays a crucial role in bacterial competition, enabling bacteria equipped with a T6SS to kill neighboring bacteria that lack this system. Additionally, some pathogens use the T6SS to deliver virulence factors into eukaryotic host cells, aiding in infection and colonization. The T6SS is tightly regulated, ensuring that it is activated only under appropriate conditions.
Gram-negative bacteria have developed specialized secretion systems to navigate their complex cellular architecture. These systems, ranging from T1SS to T4SS, are tailored to transport a variety of substrates, playing crucial roles in bacterial ecology and pathogenesis. Understanding these systems not only sheds light on bacterial physiology but also offers potential therapeutic targets against bacterial infections.
Secretion systems in Gram-negative bacteria
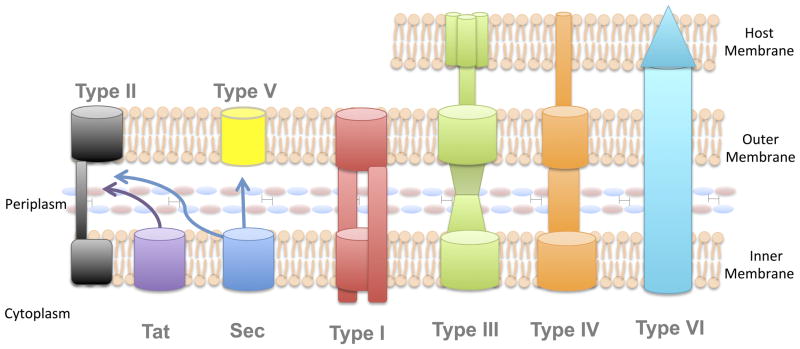
- Gram-negative bacteria have evolved intricate mechanisms to transport proteins across their complex cell envelopes, which consist of up to three phospholipid membranes. These mechanisms, known as protein secretion systems, are pivotal for bacterial survival, virulence, and interactions with their environment.
- One of the primary distinctions in these secretion systems is based on the number of steps involved in protein transport. The two-step secretion mechanisms, such as the Type II Secretion System (T2SS) and Type V Secretion System (T5SS), rely on either the Sec or Tat pathways to transport proteins across the inner membrane. Once in the periplasmic space, these proteins are then ushered out of the cell through a secondary secretion system.
- The T2SS is particularly versatile, capable of secreting proteins that were initially transported by either the Tat or Sec pathways. These proteins, once inside the periplasm, undergo folding before being secreted by the T2SS. On the other hand, the T5SS, which deals with autotransporters, requires these proteins to be in an unfolded state for transport across the outer membrane, necessitating their initial secretion via the Sec pathway.
- In contrast to the two-step systems, Gram-negative bacteria also possess one-step secretion systems, which can transport proteins directly from the bacterial cytoplasm to the external environment, bypassing the need for Sec or Tat pathways.
- These systems include the Type I, III, IV, and VI Secretion Systems (T1SS, T3SS, T4SS, and T6SS). Each of these systems is equipped with channels that span the periplasm, but their protein secretion methodologies vary significantly. Notably, the T3SS, T4SS, and T6SS are not just limited to bacterial membranes.
- They have the remarkable ability to transport proteins across the membranes of host cells, delivering their cargo directly into the host cytosol.
- In conclusion, the diverse secretion systems in Gram-negative bacteria underscore the adaptability and complexity of these microorganisms. Whether employing a one-step or two-step mechanism, these systems ensure efficient protein transport, enabling bacteria to interact with and adapt to their ever-changing environments in a myriad of ways.
Protein Secretion By Gram-positive Bacteria
Gram-positive bacteria, distinct from their Gram-negative counterparts, are characterized by a single lipid bilayer enveloped by a notably thick cell wall. Some Gram-positive species, such as Mycobacteria, even possess a lipid-rich mycomembrane. Given these structural differences, it’s logical that protein secretion mechanisms in Gram-positive bacteria differ from those in Gram-negative species.
- Basic Cellular Structure and Protein Secretion: Gram-negative bacteria have two phospholipid membranes separated by a periplasmic space with a thin peptidoglycan layer. In contrast, Gram-positive bacteria have a single lipid bilayer surrounded by a thick cell wall. This structural difference necessitates varied protein secretion mechanisms. Both Gram-positive and Gram-negative bacteria utilize the Tat and Sec pathways for protein transport across the cytoplasmic membrane. However, for Gram-positive bacteria, this transport often doesn’t suffice to deliver proteins to their ultimate destinations.
- SecA2 Secretion: The Sec secretion pathway, a highly conserved protein export mechanism, is present across bacterial classes. While it was previously believed that bacteria contained a singular SecA protein, recent findings indicate that many Gram-positive organisms, such as L. monocytogenes and M. tuberculosis, possess two SecA homologues: SecA1 and SecA2. SecA1 is essential for protein secretion via the canonical Sec pathway. In contrast, SecA2 is more specialized, often linked to stress responses or cell wall modifications.
- Sortases: Many Gram-positive pathogens express surface proteins that aid in host infection. To retain these proteins on their outer surface, Gram-positive bacteria utilize enzymes called sortases. These enzymes covalently attach proteins to the cell wall post secretion. Sortases can be general or specific in their protein targets, and they function through a transpeptidation reaction.
- Extracellular Protein Secretion: Not all proteins secreted by Gram-positive bacteria remain attached to the cell wall. Some are released into the extracellular environment, often through passive diffusion. Additionally, some Gram-positive bacteria use the Type VII Secretion System (T7SS) for protein export.
- Gram-positive “Injectosomes”: A proposed secretion apparatus in Gram-positive bacteria, termed the “injectosome,” is believed to function similarly to the T3SS and T4SS of Gram-negative bacteria. This mechanism has been observed in Streptococcus pyogenes, which injects virulence factors into host cells.
- The Type VII Secretion System (T7SS): Certain Gram-positive bacteria, especially those with a mycomembrane, utilize the T7SS for protein transport. This system is complex, with multiple components and roles in bacterial physiology and pathogenesis. For instance, while the T7SS in Listeria isn’t essential for virulence, the ESX-1 system in M. tuberculosis is crucial for its pathogenicity.
In conclusion, the protein secretion mechanisms in Gram-positive bacteria are multifaceted, influenced by their unique cellular structures. These mechanisms play pivotal roles in bacterial physiology, survival, and pathogenicity. As research progresses, a deeper understanding of these systems will undoubtedly emerge, offering potential avenues for therapeutic interventions.

Secretion systems in Gram-positive bacteria
Gram-positive bacteria are characterized by their unique cellular architecture, which includes a single cytoplasmic membrane enveloped by a robust cell wall. This structural distinction necessitates specialized mechanisms for protein secretion across the membrane. Two primary secretion systems have been identified in these bacteria: the Tat (Twin-arginine translocation) and Sec (Secretion) systems.
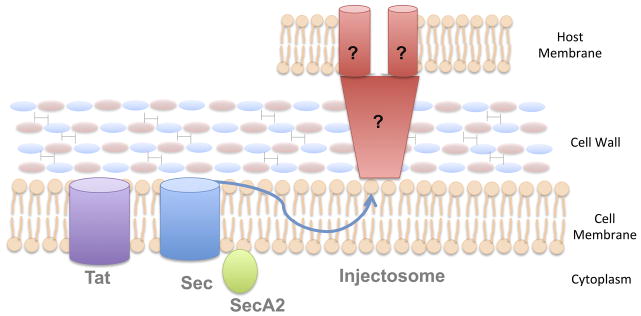
- Tat and Sec Secretion Systems: Both the Tat and Sec systems facilitate the translocation of proteins across the cytoplasmic membrane. While the Tat system is responsible for transporting fully folded proteins, the Sec system primarily secretes unfolded proteins, which are then folded outside the cell.
- SecA2 Factor: Unique to many Gram-positive bacteria is the presence of an auxiliary factor known as SecA2. This factor augments the Sec secretion system, aiding in the secretion of a specific subset of proteins. The exact role and mechanism of SecA2 remain subjects of ongoing research.
- Injectosomes: Beyond the conventional secretion systems, some Gram-positive bacteria possess specialized secretion apparatuses termed “injectosomes.” These complex structures are believed to facilitate a two-step process of protein transport. Initially, proteins are transported from the bacterial cytoplasm into the extracellular milieu. Subsequently, these proteins may be introduced into the host cell’s cytoplasm. The exact mechanism by which injectosomes achieve this dual translocation remains elusive. However, it is hypothesized that a protected channel might be employed by the injectosome to ensure safe protein transport across the thick bacterial cell wall.
In conclusion, the secretion systems in Gram-positive bacteria are intricate and multifaceted, reflecting the organisms’ adaptability and their interactions with diverse environments. Understanding these systems is pivotal for insights into bacterial physiology, pathogenesis, and potential therapeutic interventions.
Host Immune Recognition Of Bacterial Secretion Systems
- In the intricate dance between bacterial pathogens and mammalian hosts, the mechanisms by which the host immune system distinguishes between pathogenic and non-pathogenic bacteria have become a focal point of scientific inquiry. Central to this differentiation is the host’s ability to recognize specific bacterial pathogenic patterns, rather than merely identifying generic bacterial components like lipopolysaccharide (LPS) or peptidoglycan.
- A defining feature of many bacterial pathogens is their employment of specialized protein secretion systems. These systems are designed to introduce effector proteins or toxins directly into the host’s tissues or cells. Recognizing the potential threat posed by these secretion systems, the mammalian innate immune system has evolved a variety of strategies to detect their presence and activity.
- One such strategy involves the detection of bacterial products that gain access to the cytosol. Often, these are small molecules like peptidoglycan, flagellin, or LPS, which might be inadvertently introduced into the bacterial cytoplasm via secretion systems. The presence of these molecules in the bacterial cytosol can serve as a telltale sign of secretion system activity. A notable example of this detection mechanism is the Nod-like receptors (NLRs) family, which is adept at sensing cytosolic molecules such as LPS and flagellin. Upon activation, these receptors initiate a signaling cascade culminating in the release of inflammatory cytokines.
- Furthermore, the host immune system has mechanisms in place to directly detect the translocation of secreted bacterial effectors. A case in point is the ability of macrophages to detect alterations in Rho GTPases, a result of the action of the Yersinia T3SS effector YopE. This detection prompts the macrophages to initiate a response that facilitates the elimination of intracellular bacteria.
- Another intriguing aspect of the host’s defense mechanism is its ability to sense membrane disruptions caused by bacterial pore-forming proteins. These proteins, often associated with bacterial secretion systems, facilitate the delivery of effector proteins to target mammalian cells. An illustrative example is the activation of the NLRP3 receptor in response to pore formation by the SLO of Streptococcus pyogenes, leading to the production of inflammatory cytokines that aid in infection clearance.
- Lastly, emerging evidence suggests that the host immune system might be capable of detecting external components of bacterial secretion channels, such as the T3SS needle or translocon components. Recent studies have indicated that certain T3SS needle proteins, like YscF of Yersinia and MxiH from Shigella, can trigger the production of proinflammatory cytokines in host cells.
- In conclusion, the mammalian innate immune system’s ability to detect bacterial secretion systems represents a sophisticated and multi-faceted defense mechanism. This intricate interplay between host and pathogen underscores the evolutionary arms race that continues to shape the dynamics of host-pathogen interactions.
Mechanisms of innate immune recognition of bacterial secretion systems
The mammalian innate immune system, a critical component of host defense, has evolved sophisticated mechanisms to differentiate between pathogenic and commensal bacteria. Central to this discernment is the system’s ability to identify specific patterns and mechanisms associated with bacterial pathogens. One such distinctive feature is the bacterial protein secretion apparatus, which plays a pivotal role in bacterial virulence and interaction with host cells.
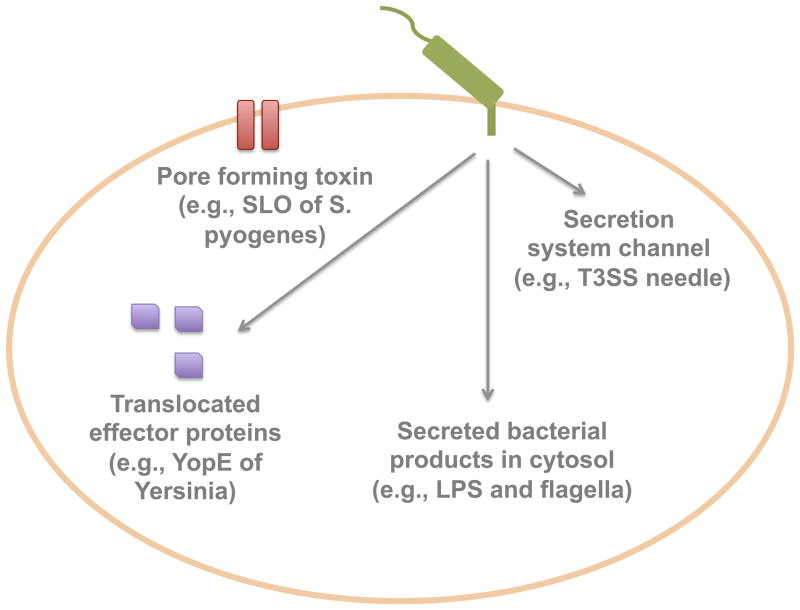
Several aspects of bacterial protein secretion are recognized by the innate immune system:
- Pore Formation Recognition: Many bacterial pathogens utilize secretion systems or specific secreted proteins that form pores in host cell membranes. The innate immune system can detect these pore formations, which often serve as entry points for bacterial effector proteins or toxins.
- Detection of Aberrant Molecule Translocation: The immune system is adept at sensing when bacterial molecules, which typically remain within the bacterial cell, are aberrantly translocated into the host cytosol. Such translocations can be indicative of bacterial secretion system activity and can trigger immune responses.
- Effector Protein Sensing: Bacterial pathogens often secrete effector proteins that manipulate host cell functions to benefit the bacteria. The presence or activity of these effector proteins can be recognized by the host immune system, leading to the initiation of defensive responses.
- Recognition of Secretion System Components: The components that constitute bacterial secretion systems, especially those that are exposed or protrude from the bacterial surface, can be direct targets for recognition by the innate immune system.
In summary, the mammalian innate immune system’s ability to detect and respond to bacterial protein secretion mechanisms underscores its evolutionary adaptability and the ongoing interplay between host and pathogen. This recognition plays a crucial role in ensuring effective host defense against bacterial invasions, maintaining a delicate balance between immune response and tolerance.
Importance of Bacterial Secretion Systems
Bacterial secretion systems play a pivotal role in the survival, adaptation, and pathogenicity of bacteria. Understanding their importance is crucial for both basic microbiology and clinical applications. Here’s a detailed overview of the significance of bacterial secretion systems:
- Pathogenicity and Virulence:
- Many bacterial pathogens utilize secretion systems to inject virulence factors directly into host cells. These factors can manipulate host cell functions, allowing the bacteria to evade the immune response, acquire nutrients, and establish infections.
- Host-Pathogen Interactions:
- Secretion systems mediate interactions between bacteria and their host organisms. They can modulate host immune responses, alter cellular functions, and even induce cell death, facilitating bacterial colonization and infection.
- Environmental Adaptation:
- Bacteria often face hostile environments, be it within a host or in external habitats. Secretion systems allow bacteria to secrete enzymes that can degrade harmful compounds or toxins, aiding in their survival.
- Biofilm Formation:
- Some secreted factors promote biofilm formation, a protective community of bacteria encased in a matrix. Biofilms enhance resistance to antibiotics and host immune responses, making infections challenging to treat.
- Nutrient Acquisition:
- Bacteria can secrete enzymes that break down complex molecules in their environment, facilitating nutrient uptake. This is especially crucial for bacteria living in nutrient-poor environments.
- Interspecies Communication:
- Secretion systems can release signaling molecules that allow communication between different bacterial species, enabling coordinated behaviors like quorum sensing.
- Antibiotic Resistance:
- Certain secretion systems can actively pump out antibiotics, contributing to antibiotic resistance. This mechanism is a significant concern in treating bacterial infections.
- Immune Evasion:
- By secreting specific proteins, bacteria can interfere with the host’s immune mechanisms, preventing recognition and clearance by the immune system.
- Cellular Adhesion:
- Some secreted factors facilitate bacterial adhesion to host cells or surfaces, a critical step in colonization and infection.
- Potential Therapeutic Targets:
- Given their essential roles in bacterial survival and virulence, secretion systems are being explored as potential targets for new antimicrobial therapies. Targeting these systems could pave the way for innovative treatments, especially against antibiotic-resistant strains.
In summary, bacterial secretion systems are multifunctional apparatuses that significantly influence bacterial behavior, interactions with their environment, and pathogenic potential. Their study not only enhances our understanding of bacterial biology but also offers avenues for therapeutic interventions.
Quiz
Which bacterial secretion system is responsible for transporting folded proteins across the cytoplasmic membrane?
a) Sec pathway
b) Tat pathway
c) T2SS
d) T3SS
The Sec pathway typically transports:
a) Folded proteins
b) Unfolded proteins
c) DNA molecules
d) RNA molecules
Which component of the Tat pathway recognizes the “twin” arginine motif on folded secretion substrates in Gram-negative bacteria?
a) TatA
b) TatB
c) TatC
d) TatD
In Gram-positive bacteria, which secretion system is used for the transport of a smaller subset of proteins?
a) SecA1
b) SecA2
c) SecB
d) SecC
Which bacterial secretion system directly deposits effector proteins or toxins into mammalian tissue sites and/or host cells?
a) T1SS
b) T2SS
c) T3SS
d) T4SS
Which of the following is NOT a component of the Sec pathway?
a) SecA
b) SecB
c) SecYEG channel
d) TatA
The Nod-like receptors (NLRs) in the mammalian immune system can directly sense:
a) LPS
b) Flagellin
c) Both LPS and Flagellin
d) Neither LPS nor Flagellin
Which secretion system in Gram-negative bacteria is responsible for exporting proteins from the periplasm to the extracellular environment?
a) T1SS
b) T2SS
c) T3SS
d) T4SS
The SRP particle in the Sec pathway recognizes proteins with a signal sequence and is involved in:
a) Post-translational mechanism
b) Co-translational mechanism
c) Both post-translational and co-translational mechanisms
d) Neither post-translational nor co-translational mechanisms
Which bacterial secretion system is known to manipulate Rho GTPases in host cells?
a) T1SS
b) T2SS
c) T3SS
d) T4SS
FAQ
What are bacterial secretion systems?
Bacterial secretion systems are specialized protein complexes that bacteria use to transport molecules, often proteins, across their cell membranes.
Why are bacterial secretion systems important?
These systems play crucial roles in bacterial survival, virulence, and interactions with host organisms. They allow bacteria to communicate, defend against hostile environments, and infect host cells.
How many types of bacterial secretion systems are there?
There are several types of bacterial secretion systems, commonly referred to as Type I through Type VII (T1SS to T7SS), each with distinct mechanisms and functions.
What is the primary function of the Sec pathway?
The Sec pathway is responsible for transporting unfolded proteins across the bacterial cytoplasmic membrane.
How does the Tat pathway differ from the Sec pathway?
Unlike the Sec pathway, which transports unfolded proteins, the Tat pathway is specialized in secreting folded proteins across the cytoplasmic membrane.
Are bacterial secretion systems present in both Gram-positive and Gram-negative bacteria?
Yes, both Gram-positive and Gram-negative bacteria possess secretion systems, but the specific systems and their mechanisms can vary between the two groups.
How do bacterial secretion systems contribute to bacterial pathogenicity?
Certain secretion systems allow bacteria to inject effector proteins directly into host cells, manipulating host cell functions and promoting bacterial survival and replication.
What role do bacterial secretion systems play in antibiotic resistance?
Some bacterial secretion systems can export antibiotics out of bacterial cells, contributing to antibiotic resistance.
Can the mammalian immune system detect bacterial secretion systems?
Yes, the mammalian immune system has evolved strategies to detect bacterial secretion systems and their secreted substrates, allowing for a rapid immune response against invading pathogens.
Are bacterial secretion systems potential targets for new antibiotics?
Yes, given their crucial role in bacterial virulence and survival, bacterial secretion systems are being studied as potential targets for the development of new antimicrobial agents.
References
- Green ER, Mecsas J. Bacterial Secretion Systems: An Overview. Microbiol Spectr. 2016 Feb;4(1):10.1128/microbiolspec.VMBF-0012-2015. doi: 10.1128/microbiolspec.VMBF-0012-2015. PMID: 26999395; PMCID: PMC4804464.
- https://www.frontiersin.org/articles/10.3389/fmicb.2022.917591
- https://www.nature.com/articles/nrmicro3456
- https://bmcmicrobiol.biomedcentral.com/articles/10.1186/1471-2180-9-S1-S2
- https://www.sciencedirect.com/topics/immunology-and-microbiology/bacterial-secretion-system
- https://www.sciencedirect.com/topics/immunology-and-microbiology/bacterial-secretion-system
- https://www.microbiologyresearch.org/content/journal/micro/10.1099/mic.0.001193?crawler=true
- https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1010610
- https://www.kegg.jp/pathway/ko03070
- https://www.kegg.jp/pathway/ko03070
- https://biomedicalsciences.unimelb.edu.au/sbs-research-groups/biochemistry-and-pharmacology-research/debnath-ghosal-laboratory/bacterial-secretion-systems
- https://link.springer.com/10.1007/978-3-540-77587-4_361


