Table of Contents
What is Genetic Linkage?
- Genetic linkage is like a genetic glue that binds DNA sequences residing close together on a chromosome, ensuring they journey together during the magical meiosis phase of sexual reproduction. It’s like friends who always stick together during a fun adventure.
- Imagine genes as signposts along a chromosome road. When two genetic markers find themselves neighbors on this road, they tend to team up during the crossing-over dance. It’s as if they hold hands and make a pact not to wander off onto different paths.
- This partnership is stronger when markers are snugly close. Genes that are far apart are more likely to part ways during this dance. In simple terms, if genes are like neighboring houses, the closer they are, the less likely they’ll move to different streets during the dance. They become linked like inseparable buddies.
- But here’s the twist: genes residing on different streets (chromosomes) aren’t linked. They can go their separate ways during the dance. However, there’s a small twist within this twist. Sometimes, certain gene versions can play mischief. They might not be on the same street but can still influence each other, a bit like friends from different neighborhoods affecting each other’s choices.
- Genetic linkage takes a step away from Gregor Mendel’s Law of Independent Assortment. Back in 1905, an experiment spilled the beans on this exceptional genetic phenomenon. At that time, scientists were puzzled about why certain traits were always packaged together. Later, they unraveled that genes are like distant cousins linked by physical distance.
- When it comes to measuring this genetic companionship, scientists use a unit called the centimorgan (cM). Imagine two markers 1 cM apart. This suggests that, on average, they split onto different paths once in every 100 meiotic productions, roughly once in every 50 dance moves.
- So, genetic linkage is like genes forming squads, holding hands, and promising to stay together on the journey of creating the next generation. It’s like a genetic partnership where distance matters and influences whether genes will stick together or go their separate ways.
Definition of Genetic Linkage
Genetic linkage is when genes situated close together on a chromosome tend to be inherited together during sexual reproduction.
Exploring Terms Related to Genetic Linkage
Genes on the Same Team: Imagine genes as friends who end up living on the same street, the chromosome. When different genes call the same chromosome home, they tend to stick together during the process of meiosis, not following the random rules. This teaming up of genes on a chromosome is called genetic linkage.
Linkage Group – The Chromosomal Gang: Think of a linkage group as a gang of genes. It’s like all the genes that hang out on a particular chromosome. These genes are physically neighbors, and they often influence each other’s inheritance patterns.
Navigating with a Linkage Map: Just as a map guides us through unknown terrain, a linkage map guides us through chromosomes. It’s like a blueprint of chromosomal locations but not based on concrete measurements. Instead, it’s crafted from the frequencies of genetic recombination, helping us understand how genes are positioned on chromosomes.
Characteristic of Linked Genes
Linked genes are genes that are located close together on the same chromosome and tend to be inherited together. Here are some key characteristics of linked genes:
- Physical Proximity: Linked genes are located close to each other on the same chromosome. Because of this proximity, they tend to not segregate independently during meiosis.
- Reduced Recombination: Linked genes have a lower chance of being separated by recombination during meiosis compared to genes that are farther apart. This is because the likelihood of a crossover event occurring between them is reduced.
- Inheritance Patterns: Linked genes often show non-Mendelian inheritance patterns. Instead of the expected 9:3:3:1 ratio seen in dihybrid crosses with independently assorting genes, linked genes might show ratios like 9:3:4 or other variations.
- Recombination Frequency: The probability that a crossover will occur between two linked genes can be used to estimate the distance between them on a chromosome. This is measured in centimorgans (cM). A recombination frequency of 1% is equivalent to a distance of 1 cM.
- Genetic Maps: Using the recombination frequencies between different pairs of genes, geneticists can create genetic maps that show the relative positions of genes on a chromosome.
- Coupling and Repulsion: When studying linked genes, two types of heterozygous gene arrangements can be observed:
- Coupling (cis configuration): Where the dominant alleles of two genes are on one chromosome and the recessive alleles are on the other chromosome.
- Repulsion (trans configuration): Where one dominant allele and one recessive allele of the two genes are found on the same chromosome.
- Incomplete Linkage: No two genes are completely linked, meaning that there is always some chance of recombination between them. The degree of linkage between two genes can vary, with some being more tightly linked than others.
- Discovery: The concept of gene linkage was first introduced by Thomas Hunt Morgan and his student Alfred Sturtevant while studying fruit flies. They noticed that certain traits were often inherited together, leading to the discovery of linked genes.
- Importance in Evolution: Linkage can slow down the process of genetic variation since linked genes are inherited together. However, recombination events can introduce new gene combinations, providing variation for natural selection to act upon.
- Disease Association: Sometimes, multiple genes that are linked can be associated with a particular disease or condition. This can be useful in genetic studies to identify regions of the genome associated with specific diseases.
Understanding gene linkage is crucial in genetics, especially in the fields of genetic mapping, breeding, and evolutionary biology.
What is Linkage map?
A linkage map, commonly referred to as a genetic map, is a structured representation that delineates the relative positions of genes or genetic markers on a chromosome. This positioning is not based on the precise physical locations on the chromosome but rather on the recombination frequency between these genes or markers. The foundational principle behind linkage maps is that the probability of recombination between two genetic loci is proportional to the distance separating them.
- Historical Context: The concept of the linkage map was pioneered by Alfred Sturtevant, a student under the tutelage of the eminent geneticist Thomas Hunt Morgan. Sturtevant’s groundbreaking work laid the groundwork for subsequent genetic research, emphasizing the importance of understanding gene positions relative to one another.
- Mechanistic Underpinning: The essence of a linkage map is rooted in the phenomenon of crossing over, which occurs during the prophase of meiosis. During this process, homologous non-sister chromatids undergo an exchange of chromosomal segments. The frequency of such recombination events between two markers serves as an indirect measure of their relative distance. A higher recombination frequency indicates that the markers are farther apart, while a lower frequency suggests proximity.
- Markers in Linkage Maps: Historically, the markers employed in linkage maps were discernible phenotypic traits, such as eye color or enzyme production, which were manifestations of coding DNA sequences. With advancements in molecular genetics, researchers have transitioned to using noncoding DNA sequences, like microsatellites or restriction fragment length polymorphisms (RFLPs), as markers.
- Applications and Utility: Linkage maps serve as invaluable tools for geneticists, facilitating the identification and localization of new genes or markers. Initially, data from linkage maps are employed to establish linkage groups, which are clusters of genes known to be linked. As research progresses, these groups can be expanded to encompass an entire chromosome. For well-researched organisms, these linkage groups correspond directly to individual chromosomes.
- Distinguishing from Other Maps: It is imperative to differentiate linkage maps from physical or gene maps. While linkage maps are predicated on recombination frequencies, physical maps provide a detailed representation of the actual physical distances between genes or markers on a chromosome.
Example from Scientific Literature: In a seminal research paper titled “Isozymic gene linkage map of the tomato: Applications in genetics and breeding” by Steven D. Tanksley and Charles M. Rick, the authors elucidated the linkage map of the tomato, emphasizing its implications in genetic studies and breeding practices1. Such research underscores the significance of linkage maps in understanding the genetic architecture of organisms and their subsequent applications in breeding and genetic improvement.
In conclusion, linkage maps are indispensable tools in the realm of genetics, offering insights into the relative positions of genes and facilitating a deeper understanding of the genetic landscape of organisms.
Chromosome Theory of Linkage
The Chromosome Theory of Linkage is an extension of the Chromosome Theory of Inheritance. It provides an explanation for the way in which genes are transmitted through generations in a linked manner. Here’s a breakdown of the theory:
- Basis in Chromosome Theory of Inheritance: The Chromosome Theory of Inheritance, proposed by Walter Sutton and Theodor Boveri, posits that genes are located on chromosomes, and it is the chromosomes that segregate and assort independently during meiosis, leading to the inheritance of traits. The Chromosome Theory of Linkage builds upon this by explaining how genes on the same chromosome can be inherited together.
- Genes on the Same Chromosome: The central idea of the Chromosome Theory of Linkage is that genes located close together on the same chromosome are usually inherited together. This is because they do not assort independently like genes on different chromosomes.
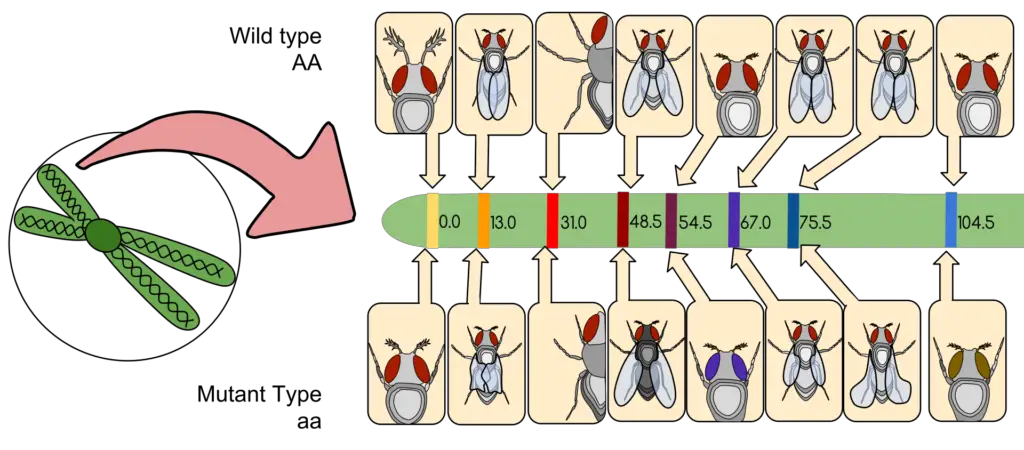
- Discovery by Morgan’s Work: Thomas Hunt Morgan, working with fruit flies (Drosophila melanogaster), provided experimental evidence supporting the Chromosome Theory of Linkage. He observed that certain traits, which should have assorted independently according to Mendelian genetics, did not. This led him to propose that these genes were linked on the same chromosome.
- Recombination and Crossing Over: While genes on the same chromosome are often inherited together, they are not always. This is due to the process of crossing over during meiosis. In crossing over, homologous chromosomes exchange segments, leading to a recombination of genes. The frequency of recombination between two genes is proportional to the distance between them on the chromosome.
- Genetic Mapping: Using the frequency of recombination, Alfred Sturtevant, a student of Morgan, was able to create the first genetic map. He proposed that the farther apart two genes are on a chromosome, the higher the chance of a crossover event occurring between them, and thus, the higher the recombination frequency. This led to the concept of map units or centimorgans to measure genetic distance.
- Variation in Linkage: Not all genes on the same chromosome are linked to the same degree. The strength of linkage between two genes is determined by the distance between them. Genes that are closer together are more strongly linked and have a lower recombination frequency.
- Importance in Evolution and Breeding: Understanding linkage is crucial for breeders and evolutionary biologists. For breeders, linkage can be both an advantage and a challenge, as desired traits might be linked to undesired ones. For evolutionary biologists, linkage can affect the rate at which new genetic combinations arise in populations.
In summary, the Chromosome Theory of Linkage provides a framework for understanding how genes on the same chromosome can be inherited together, the role of recombination in generating genetic diversity, and the importance of linkage in genetics and evolution.
Types of Linkage
In the realm of genetics, the pioneering work of Thomas Hunt Morgan and his associates with the fruit fly Drosophila yielded crucial insights into the complexities of gene inheritance. Their research led to the identification of two fundamental types of linkage – complete linkage and incomplete linkage – shedding light on the diverse ways in which genes are inherited together or independently.
1. Complete Linkage
Complete linkage refers to the situation where two genes are located so close together on the same chromosome that they are always inherited together and do not show any recombination. In other words, the genes do not assort independently, and there is no crossing over between them during meiosis. As a result, the offspring inherit the same combinations of alleles as the parents.
Example of Complete Linkage:
Let’s consider a hypothetical example involving two genes in a plant:
- Gene A determines flower color:
- A allele results in red flowers.
- a allele results in white flowers.
- Gene B determines leaf shape:
- B allele results in broad leaves.
- b allele results in narrow leaves.
Suppose these two genes are located very close together on the same chromosome, leading to complete linkage.
Now, let’s cross two heterozygous plants (AaBb). If these genes were assorting independently (following Mendel’s law of independent assortment), we would expect a 9:3:3:1 phenotypic ratio in the offspring:
- 9 red flowers with broad leaves (A-B-)
- 3 red flowers with narrow leaves (A-bb)
- 3 white flowers with broad leaves (aaB-)
- 1 white flower with narrow leaves (aabb)
However, due to complete linkage, the genes will always be inherited together. So, when crossing two heterozygous plants (AaBb x AaBb), the expected offspring would be:
- Red flowers with broad leaves (from AB gametes)
- White flowers with narrow leaves (from ab gametes)
There would be no offspring with red flowers and narrow leaves or white flowers and broad leaves because there’s no recombination between the genes.
In reality, complete linkage is rare, as even genes that are very close together on a chromosome can still undergo crossing over, albeit at a very low frequency. Most linked genes show some degree of recombination, leading to incomplete linkage.
2. Incomplete Linkage
Incomplete linkage refers to the phenomenon where linked genes, which are located relatively far apart on the same chromosome, do not always inherit together due to the possibility of being separated by crossing over. While these genes are on the same chromosome and tend to be inherited together more often than not, they can still be separated during meiosis.
The reason for this separation is that during the prophase of meiosis, homologous non-sister chromatids can exchange segments of varying lengths. This process, where segments of chromosomes are swapped between homologous pairs, is termed “crossing over.” As a result of crossing over, new combinations of alleles can be produced, leading to genetic variation.
Example of Incomplete Linkage:
One notable example of incomplete linkage is observed in maize (Zea mays). This was studied by Hutchison, who looked at the inheritance of two traits: the color and shape of the seed.
- Initial Cross:
- A maize plant with seeds that are colored and have a full endosperm (represented by the genotype CS/CS) is crossed with another plant that has seeds which are colorless and shrunken (genotype cs/cs).
- F1 Generation:
- The resulting F1 heterozygotes from this cross display seeds that are colored and full, with a genotype of CS/cs.
- Test Cross:
- When these F1 hybrids are test crossed with the double recessive parent (cs/cs), an interesting pattern emerges. Instead of obtaining just two classes of offspring (which would be expected with complete linkage), four distinct classes are observed.
- Results:
- 96% of the offspring display the parental combinations of alleles, either CS/cs or cs/cs. This high percentage indicates that the genes are linked and tend to be inherited together.
- However, 4% of the offspring show new combinations of alleles, either Cs/cs or cS/cs. These new combinations are a direct result of crossing over between the linked genes during meiosis.
This example in maize clearly demonstrates that while the genes for seed color and shape are linked, they do not show complete linkage. The presence of new allele combinations in 4% of the offspring is evidence of crossing over, highlighting the phenomenon of incomplete linkage.
Furthermore, it’s worth noting that incomplete linkage has been observed in various organisms, including female Drosophila, tomato, pea, mice, poultry, and humans.
In conclusion, Morgan’s groundbreaking work with Drosophila unveiled the existence of two distinct types of linkage – complete and incomplete – each manifesting different patterns of gene inheritance. These findings not only solidified the chromosome theory of inheritance but also provided a deeper understanding of the mechanisms underlying trait transmission. The concepts of complete and incomplete linkage underscore the intricate interplay between gene location on chromosomes, genetic recombination, and the observed inheritance patterns that form the foundation of modern genetics.
Linkage analysis
Linkage analysis is a sophisticated genetic technique employed to identify chromosomal segments that consistently co-segregate with a specific phenotype across multiple generations within families. This method has been pivotal in mapping genes associated with both binary (dichotomous) and quantitative (continuous) traits.
Types of Linkage Analysis
- Parametric Linkage Analysis: This approach necessitates prior knowledge regarding the relationship between phenotypic and genetic similarity. The cornerstone of parametric linkage analysis is the LOD (logarithm of odds) score. The LOD score is a statistical measure that evaluates the likelihood of observing the test data under the assumption of linkage between two loci, compared to the likelihood of observing the same data by mere coincidence. A positive LOD score suggests the presence of linkage, while a negative score implies that linkage is improbable.
- Non-parametric Linkage Analysis: This method does not rely on prior assumptions about the genetic model. Instead, it evaluates the probability of an allele being identical by descent.
LOD Score Computation
The LOD score, a brainchild of Newton Morton, is computed using the formula:
LOD = Z =
Where:
- NR represents the number of non-recombinant offspring.
- R represents the number of recombinant offspring.
- θ is the recombination fraction, signifying the proportion of offspring in which recombination occurs between the genetic marker under study and the hypothetical gene associated with the disease.
Applications
Linkage analysis facilitates the identification of genetic markers in close proximity to genes that influence specific traits or diseases. For instance, through linkage analysis, researchers can pinpoint genes associated with rare genetic disorders.
Limitations
Despite its utility, linkage analysis is not devoid of limitations. It possesses both methodological and theoretical constraints that can elevate the type-1 error rate and diminish the power to map quantitative trait loci (QTL) in humans. While linkage analysis has been instrumental in identifying genetic variants linked to rare disorders, its efficacy diminishes when applied to prevalent disorders like cardiovascular diseases or various cancers. This discrepancy can be attributed to the distinct genetic underpinnings of common versus rare disorders.
Example
Consider a pedigree where a specific genetic disorder is prevalent. Through parametric linkage analysis, researchers can assess the probability that a particular genetic marker and the disease gene are linked based on the observed patterns of inheritance within the family. If the LOD score exceeds the threshold of 3.0, it provides compelling evidence for linkage, suggesting that the marker and the disease gene are located in close proximity on the chromosome.
In summary, linkage analysis is a robust genetic tool that has significantly advanced our understanding of the genetic architecture of various traits and diseases. Its precision and methodological rigor make it an indispensable technique in the realm of genetic research.
Importance of Linkage
The significance of linkage can be elucidated as follows:
- Genetic Mapping: Linkage serves as the foundational principle for constructing genetic maps. The frequency of recombination between genes, which is inversely proportional to the distance between them on a chromosome, provides a metric to determine their relative positions. This has been instrumental in mapping the human genome and those of numerous other organisms.Example: Alfred Sturtevant’s pioneering work on Drosophila melanogaster led to the formulation of the first genetic map, where he used recombination frequencies to deduce the relative positions of genes on the chromosome.
- Genetic Variation and Evolution: While linkage can constrain genetic variation due to the co-inheritance of linked alleles, crossing over introduces new gene combinations. This balance between linkage and recombination ensures a steady influx of genetic variation, which is vital for populations to adapt and evolve in changing environments.Example: In populations exposed to fluctuating environmental conditions, the interplay between linkage and recombination can lead to the emergence of adaptive gene combinations that confer a selective advantage.
- Genetic Disorders and Linkage Disequilibrium: Linkage disequilibrium (LD) refers to the non-random association of alleles at different loci. LD is a crucial concept in the study of genetic disorders, as certain combinations of alleles can predispose individuals to specific diseases.Example: The study of LD patterns has been pivotal in identifying genetic markers associated with complex diseases like Alzheimer’s, where specific allele combinations increase disease susceptibility.
- Breeding and Agriculture: In crop breeding, understanding linkage is paramount. Desirable traits that are linked can be co-selected, but linkage can also pose challenges when beneficial traits are linked to undesirable ones.Example: In wheat breeding, a desired trait for disease resistance might be linked to an undesired trait affecting grain quality. Breeders must employ strategies like marker-assisted selection to break this linkage and achieve the desired genetic combination.
- Molecular Evolution and Genomic Studies: Linkage can influence patterns of molecular evolution. Genes within tightly linked regions might experience reduced effective population sizes, affecting their susceptibility to genetic drift and selective pressures.Example: In genomic scans for selection, regions of reduced genetic variation might indicate selective sweeps, where beneficial mutations and linked neutral mutations increase in frequency together.
In conclusion, the phenomenon of linkage, while seemingly a straightforward genetic concept, has far-reaching implications in diverse fields, from molecular genetics to evolutionary biology and agriculture. Understanding the intricacies of linkage and its consequences is paramount for advancements in genetic research and its applications.
Genetic Linkage Mindmap

FAQ
What is genetic linkage?
Genetic linkage refers to the tendency of genes located close together on the same chromosome to be inherited together more often than expected by chance.
How was genetic linkage discovered?
Genetic linkage was first observed by Thomas Hunt Morgan and his student Alfred Sturtevant while studying fruit flies (Drosophila melanogaster). They noticed that certain genes did not assort independently, as predicted by Mendel’s laws.
What is a linkage map?
A linkage map, or genetic map, is a representation that shows the relative positions of genes or genetic markers on a chromosome based on recombination frequencies, rather than actual physical distances.
How does crossing over relate to genetic linkage?
Crossing over is the process where homologous chromosomes exchange segments during meiosis. This can separate genes that are linked, leading to recombination. The frequency of crossing over between two genes is inversely proportional to their linkage strength.
What is a LOD score in linkage analysis?
The LOD (logarithm of odds) score is a statistical measure used in linkage analysis to determine the likelihood of two genes being linked. A LOD score greater than 3 typically indicates significant evidence for linkage.
Why are some genes more tightly linked than others?
The strength of linkage between two genes is determined by their distance apart on a chromosome. Genes that are closer together are more tightly linked and have a lower recombination frequency.
What is the difference between complete linkage and incomplete linkage?
Complete linkage occurs when genes are so closely located on a chromosome that they are always inherited together. Incomplete linkage, on the other hand, refers to genes that are linked but can still be separated by crossing over due to their relative distance on the chromosome.
How do linkage maps aid in genetic research?
Linkage maps allow researchers to determine the relative positions of genes on a chromosome. This can help in identifying genes responsible for certain traits or diseases and is crucial for breeding programs and evolutionary studies.
What is linkage disequilibrium?
Linkage disequilibrium (LD) refers to the non-random association of alleles at different loci. It indicates that the alleles at two linked loci occur together more or less frequently than would be expected by chance.
Can genes on different chromosomes be linked?
No, linkage specifically refers to genes on the same chromosome. However, genes on different chromosomes can show non-random associations due to other factors, but this is not considered genetic linkage.
References
- Tanksley, S.D., & Rick, C.M. (1980). Isozymic gene linkage map of the tomato: Applications in genetics and breeding.
- https://www.genome.gov/genetics-glossary/Linkage
- https://learn.genetics.utah.edu/content/pigeons/geneticlinkage/
- https://www.khanacademy.org/science/ap-biology/heredity/non-mendelian-genetics/a/linkage-mapping
- http://www.nature.com/scitable/topicpage/discovery-and-types-of-genetic-linkage-500
- https://bio.libretexts.org/Bookshelves/Introductory_and_General_Biology/Introductory_Biology_(CK-12)/03%3A_Genetics/3.10%3A_Genetic_Linkage
- https://www.nature.com/articles/nrg3908
- https://www.sciencedirect.com/topics/agricultural-and-biological-sciences/genetic-linkage
- https://pubmed.ncbi.nlm.nih.gov/10369304/


