Table of Contents
What is Post-transcriptional Modification?
- Post-transcriptional modification refers to the various changes and alterations that occur to RNA molecules after transcription, the process by which RNA is synthesized from DNA. These modifications play crucial roles in regulating gene expression and expanding the functional diversity of RNA molecules.
- After transcription, primary RNA transcripts, known as pre-mRNA, undergo several modifications to produce mature mRNA that can be translated into proteins. Some of the common post-transcriptional modifications include alternative splicing, capping, polyadenylation, and RNA editing.
- Alternative splicing is a process where different combinations of exons within a pre-mRNA are selected, leading to the production of multiple mRNA isoforms from a single gene. By selectively including or excluding certain exons, alternative splicing allows for the generation of different protein variants with distinct functions. This process greatly increases the proteome diversity and contributes to the complexity of higher organisms.
- Capping involves the addition of a modified guanine nucleotide (the 5′ cap) to the 5′ end of the pre-mRNA molecule. This cap structure provides stability to the RNA molecule and is essential for efficient translation. It also plays a role in regulating mRNA processing, transport, and degradation.
- Polyadenylation is the addition of a poly(A) tail to the 3′ end of the mRNA molecule. The poly(A) tail consists of a string of adenine nucleotides and serves multiple functions. It helps protect the mRNA from degradation, assists in the export of mRNA from the nucleus to the cytoplasm, and plays a role in translation initiation and mRNA stability.
- RNA editing is a process where specific nucleotides within the RNA sequence are enzymatically modified. This can involve the conversion of adenosine (A) to inosine (I) or the deamination of cytosine (C) to uracil (U). RNA editing can alter the coding potential of mRNA, leading to changes in the amino acid sequence of the resulting protein.
- These post-transcriptional modifications are highly regulated and can influence gene expression patterns and protein diversity in various ways. They allow cells to fine-tune gene expression levels, produce different protein isoforms with specialized functions, and respond to environmental cues or cellular signals. Dysregulation of post-transcriptional modifications can have profound effects on cellular processes and contribute to the development of diseases such as cancer and neurological disorders.
- To study post-transcriptional modifications, researchers employ various techniques such as high-throughput sequencing, RNA immunoprecipitation, and bioinformatics analysis. These methods enable the identification and characterization of modified RNA molecules, providing insights into their functional significance and regulatory mechanisms.
- In summary, post-transcriptional modification encompasses a range of processes that modify RNA molecules after transcription. These modifications contribute to the complexity and diversity of the proteome by regulating gene expression and expanding the functional repertoire of RNA molecules. Understanding post-transcriptional modifications is essential for unraveling the intricacies of gene regulation and its impact on cellular function and human health.
Definition of Post-transcriptional Modification
Post-transcriptional modification refers to the chemical alterations and changes that occur to RNA molecules after they have been transcribed from DNA. These modifications can include processes such as alternative splicing, capping, polyadenylation, and RNA editing, which collectively regulate gene expression and contribute to the diversity of proteins produced in a cell.
Location of Post-transcriptional Modification
- Post-transcriptional modifications occur in the nucleus and cytoplasm of eukaryotic cells. The primary site of post-transcriptional modifications is the nucleus, where processes like splicing, capping, and polyadenylation take place. Splicing, the removal of introns and joining of exons, occurs within the nucleus. Capping, the addition of a modified guanine nucleotide to the 5′ end of mRNA, also occurs in the nucleus. Polyadenylation, the addition of a poly(A) tail to the 3′ end of mRNA, primarily takes place in the nucleus.
- After these initial modifications, the mature mRNA molecules are transported from the nucleus to the cytoplasm, where they undergo additional post-transcriptional modifications and participate in translation. RNA editing, which involves specific nucleotide modifications, can occur in the nucleus or cytoplasm, depending on the type of editing.
- In prokaryotic cells, which lack a nucleus, post-transcriptional modifications occur in the cytoplasm. Processes such as mRNA processing and modification, including cleavage, polyadenylation, and RNA editing, take place directly in the cytoplasm.
- Overall, the location of post-transcriptional modifications varies depending on the cell type and the specific modification being carried out.
Importance of Post-transcriptional modifications
Post-transcriptional modifications play a crucial role in the regulation and efficiency of protein synthesis, thereby impacting various cellular processes. Here are some key reasons highlighting the importance of post-transcriptional modifications:
- mRNA Stability and Degradation: Post-transcriptional modifications, such as the addition of a 5′ cap and a poly(A) tail, enhance the stability of mRNA molecules. These modifications protect mRNA from degradation by exonucleases, ensuring a longer lifespan for mRNA in the cytoplasm. Additionally, specific modifications can mark mRNA for degradation, enabling the control of gene expression levels.
- mRNA Localization and Transport: Certain post-transcriptional modifications facilitate the localization and transport of mRNA molecules within the cell. These modifications can include the addition of RNA-binding proteins or specific RNA sequences that mediate mRNA transport to distinct subcellular compartments. This localization allows mRNA to be translated near its site of function, improving the efficiency of protein synthesis and local cellular responses.
- Alternative Splicing and Protein Diversity: Alternative splicing, a post-transcriptional modification, allows the generation of multiple mRNA isoforms from a single gene. By selectively including or excluding exons, alternative splicing leads to the production of different protein variants with distinct functions. This process significantly expands the protein diversity within an organism, enabling the fine-tuning of cellular processes and contributing to the complexity of higher organisms.
- Regulation of Translation Initiation: Post-transcriptional modifications can influence translation initiation, the process by which ribosomes bind to mRNA and begin protein synthesis. Modifications such as the 5′ cap and certain RNA-binding proteins facilitate the recognition of mRNA by translation initiation factors, promoting efficient ribosome binding and translation. These modifications ensure precise control over the translation of specific mRNA molecules, thereby regulating gene expression.
- RNA Editing and Protein Function: RNA editing, a post-transcriptional modification, involves altering specific nucleotides within the RNA sequence. This modification can result in changes to the amino acid sequence of the translated protein. RNA editing plays a crucial role in expanding the functional diversity of proteins and can impact protein function, stability, and interactions. It provides an additional layer of regulation and adaptability in gene expression.
Overall, post-transcriptional modifications are essential for optimizing protein synthesis, regulating gene expression, and expanding the functional repertoire of proteins. They ensure the accurate processing, stability, and efficient translation of mRNA molecules, allowing cells to finely tune their gene expression patterns and respond to dynamic environmental cues. Without these modifications, protein synthesis would be less precise, and the complexity of cellular processes would be compromised.
Types of post-translational modification
Based on the addition of chemical groups
Post-translational modifications (PTMs) based on the addition of chemical groups are diverse and have significant impacts on protein structure, function, and cellular processes. Here are some common types of post-translational modifications based on the addition of chemical groups:
- Phosphorylation: Phosphorylation is one of the most prevalent post-translational modifications and involves the addition of a phosphate group to specific amino acid residues, such as serine, threonine, or tyrosine. This modification is catalyzed by protein kinases and can regulate protein activity, cellular signaling pathways, and protein-protein interactions. Phosphorylation is crucial for processes like cell cycle regulation, signal transduction, and enzyme activation/inactivation.
- Acetylation: Acetylation is the addition of an acetyl group to the amino group of lysine residues. It is mediated by acetyltransferase enzymes and can impact protein stability, protein-protein interactions, DNA-binding ability, and gene expression regulation. Acetylation often occurs in the context of histones and is involved in chromatin remodeling and epigenetic regulation. It also plays a role in the modulation of transcription factors and the regulation of metabolic enzymes.
- Hydroxylation: Hydroxylation involves the addition of a hydroxyl group (-OH) to specific amino acid residues, such as proline or lysine. This modification is catalyzed by hydroxylases and is important for the stability and structure of certain proteins. For example, hydroxylation of proline residues is critical for collagen formation and stability. Hydroxylation of other amino acids, like lysine, can affect protein-protein interactions, protein folding, and enzymatic activity.
- Methylation: Methylation is the addition of a methyl group (-CH3) to specific amino acid residues, typically lysine or arginine. This modification is catalyzed by methyltransferase enzymes and plays diverse roles in protein function and gene expression regulation. Methylation can influence protein-protein interactions, gene transcription, chromatin structure, and signal transduction pathways. It is involved in processes like epigenetic regulation, histone modification, and the control of protein localization and stability.
Based on the addition of complex groups
- Glycosylation: Glycosylation entails the attachment of an oligosaccharide referred to as a ‘glycan’ to either a nitrogen atom (N-linked glycosylation) or an oxygen atom (O-linked glycosylation). N-linked glycosylation occurs on the asparagine amide nitrogen, whereas O-linked glycosylation occurs on the serine or threonine oxygen atom. On the surface of cells are carbohydratess in the form of N-linked or O-linked oligosaccharides that secrete proteins. They play crucial functions in protein classification, immune recognition, receptor binding, inflammation, and pathogenicity. N-linked glycans on an immune cell, for instance, can dictate its migration to specific locations. Likewise, it can determine how a cell recognizes’self’ and ‘non-self’.
- AMPylation: Protein AMPylation is the reversible addition of AMP. A phosphodiester bond is formed between the hydroxyl group of the protein and the phosphate group of AMP.
- Lipidation: lipidation is the covalent attachment of a lipid group to a protein. Prenylation, N-myristoylation, palmitoylation, and glycosylphosphatidylinositol (GPI)-anchor addition are subcategories of lipidation. Prenylation entails the addition of an isoprenoid moiety to a substrate protein’s cysteine residue. It plays a crucial role in regulating the localization and activity of several proteins with essential regulatory functions. Myristoylation entails the formation of an amide bond between a myristoyl group and a glycine residue. It is necessary for membrane association and apoptosis. A palmitoyl group is added to a cysteine residue of a protein during palmitoylation. The protein’s carboxy-terminal signal peptide is split and substituted by a GPI anchor during GPI-anchor addition. Recent studies in human genetics have shown that GPI anchors are essential to human health. Any defects in the assembly, attachment, or remodelling of GPI anchors result in inherited GPI deficiency diseases.
Based on the addition of polypeptides – Ubiquitination
Ubiquitination
- Ubiquitination entails the addition of ubiquitin, an abundant protein, to the lysine residue of a substrate. Attachment of a single ubiquitin molecule (monoubiquitination) or a chain of multiple ubiquitin molecules (polyubiquitination) is possible.
- The 26S proteasome recognises polyubiquitinated proteins and subsequently targets them for proteolysis or degradation. Monoubiquitinated proteins may have an effect on cell localization and endocytosis.
Based on the cleavage of proteins – Proteolysis
Proteolysis
Proteolysis is the process by which proteins are broken down into smaller polypeptides or amino acids. For instance, removal of N-terminal methionine, a signal peptide, after translation results in the activation of an inactive or nonfunctional protein.
Based on the amino acid modification – Deamidation
Deamidation
Deamidation is the elimination or transformation of an asparagine or glutamine residue into a different functional group. Asparagine is converted to aspartic acid or isoaspartic acid, and glutamine to glutamic acid or pyroglutamic acid. This modification can alter the structure, stability, and function of the protein.
Post-transcriptional modification Steps
The following modifications occur during the Post-transcriptional modification of proteins;
Addition of 5′ cap
- The ends of eukaryotic transcript are processed. Modified guanosine, 7- methyl guanosine is added to the 5′ end in an uncommon 5′-to-5′ (instead of 3′-to-5′) linkage; this terminal group is known as a cap.
- After completion of transcription, the 5′ end of the RNA transcript carries a free triphosphate group since it was the first incorporated nucleotide in the chain. The capping process replaces the triphosphate group with another structure called the “cap”. The cap is added by the enzyme guanyl transferase. This enzyme catalyzes the reaction between the 5′ end of the RNA transcript and a guanine triphosphate (GTP) molecule.
- During the reaction, the beta phosphate of the RNA transcript displaces a pyrophosphate group at the 5′ position of the GTP molecule. The cap is formed through a 5′-5′ linkage between the two substrates .
- Capping protects the 5’ from enzymatic degradation in the nucleus and assists in export to the cytosol.
- Eukaryotic m RNAs lacking the cap are not efficiently translated.
Process of 5′ capping
This capping process involves the addition of an extra nucleotide at the 5′ end of the mRNA and methylation by the addition of a methyl group (CH3) to the base in the newly added neucleotide and (may be) to the 2’–OH group of the sugar of one or more nucleotides at the 5′ end.
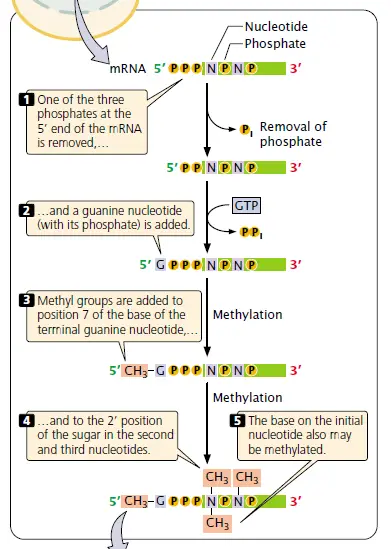
For easy understanding:
- Addition of an extra nucleotide at the 5′ end of the mRNA.
- Methylation to the base (at position 7. so, 7-methyl) in the newly added neucleotide.
- Methylation (may be) to the 2’–OH group of the sugar of one or more nucleotides at the 5′ end (second and third Sugars in the figure below).
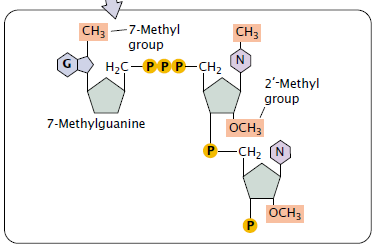
Importance of 5’ capping
- It is necessary for the mRNA to bind with the ribosome to begin protein synthesis (Cap binding proteins first identify the cap and attach to it; a ribosome then binds to these proteins and moves downstream along the mRNA until the start codon is reached and translation begins).
- Its presence also increases the stability of mRNA and influences the removal of introns.
The Addition of the Poly A tail
- The 3′ terminus of a eukaryotic mRNA molecule is transformed by the addition of a polyadenosine sequence (the poly-A tail) of as many as 50 to 250 nucleotides. These nucleotides are not encoded in the DNA but are added after transcription in a process known as poly-adenylation.
- Post-transcriptional RNA processing at the opposite end of the transcript comes in the form of a string of adenine bases attached to the end of the synthesized RNA chain.
- The addition of the adenines is catalyzed by the enzyme poly (A) polymerase.
- The mRNA is first cleaved about 20 nucleotides downstream from an AAUAA recognition sequence.
- Another enzyme, poly(A) polymerase, adds a poly(A) tail which is subsequently extended to as many as 200 A residues.
- The poly(A) tail appears to protect the 3′ end of mRNA from 3′ 5′ exonuclease attack.
- Histone and interferon’s mRNAs lack poly A tail.
- After the m-RNA enters the cytosol, the poly A tail is gradually shortened.
Process of addition
- Processing of the 3′ end of pre-mRNA requires sequences both upstream and downstream of the cleavage site.
- The consensus sequence AAUAAA is usually from 11 to 30 nucleotides upstream of the cleavage site and determines the point at which cleavage will take place.
- A sequence rich in Us (or Gs and Us) is typically downstream of the cleavage site.
- At the cleavage site, as many as 250 nucleotides are added following a complex process with the help some factors.
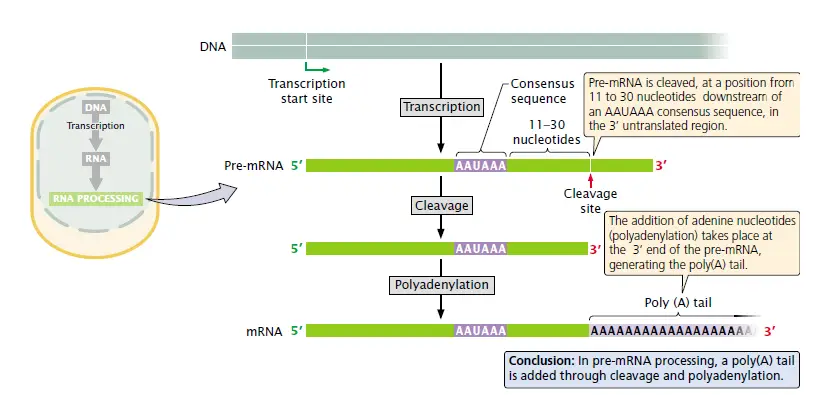
Importance of Poly A tail
- The poly(A) tail confers stability on many mRNAs, increasing the time during which the mRNA remains intact and available for translation before it is degraded by cellular enzymes.
RNA Splicing
RNA splicing is the process by which introns, regions of RNA that do not code for proteins, are removed from the pre-mRNA and the remaining exons connected to re-form a single continuous molecule.
- Splicing occurs in the nucleus following transcription but before the RNA moves to the cytoplasm.
- RNA splicing takes place in nuclear particles known as spliceosomes. These abundant particles are composed of protein and several types of specialized small nuclear RNA (snRNA) molecules ranging from 100 to 200 bases in length.
- Introns or intervening sequences are the RNA sequences which do not code for the proteins.
- These introns are removed from the primary transcript in the nucleus, exons (coding sequences) are ligated to form the mRNA molecule, and the mRNA molecule is transported to the cytoplasm.
- The molecular machine that accomplishes the task of splicing is known as the spliceosome.
- Small nuclear RNA molecules that recognize splice sites in the pre- mRNA sequence.
- The excised intron is released as a “lariat” structure, which is degraded.
- The exons portion of mRNA translated into a protein. These are the coding portions of a mRNA molecule.
- RNA splicing mainly occurs after the complete synthesis and end-capping of the pre-mRNA, transcripts with many exons can be spliced co-transcriptionally.
- This reaction is catalyzed by a large protein complex known as spliceosome, which is consists of proteins and small nuclear RNA molecules that recognize splice sites in the pre-mRNA sequence.
- Many pre-mRNAs, including those encoding antibodies, can be spliced in multiple ways to create different mature mRNAs that encode different protein sequences. This process is known as alternative splicing, and allows production of a large variety of proteins from a limited amount of DNA.
Alternative Splicing
- Alternative splicing, or alternative RNA splicing, or differential splicing, is an alternative splicing process during gene expression that allows a single gene to code for multiple proteins.
- In this process, particular exons of a gene may be included within or excluded from the final, processed messenger RNA (mRNA) produced from that gene. This means the exons are joined in different combinations, leading to different (alternative) mRNA strands.
- The proteins translated from alternatively spliced mRNAs will contain differences in their amino acid sequence and, often, in their biological functions.
- Alternative patterns of RNA splicing is adapted for the synthesis of tissue-specific proteins.(antibodies).
- The pre-mRNA molecules from some genes can be spliced in two or more alternative ways in different tissues.
- This produces multiple variations of the mRNA and thus diverse sets of proteins can be synthesized from a given set of genes.
- Alternative splicing occurs as a normal phenomenon in eukaryotes.
- Introns are removed from the primary transcript in the nucleus, exons (coding sequences) are ligated to form the mRNA molecule. (After removal of all the introns, the mature m RNA molecules leave the nucleus by passing in to the cytosol through pores in to the nuclear membrane).
- Alternative splicing accomplishes this in two ways: a) by splicing together exons from two different primary RNA transcripts in a process called trans- splicing b) by splicing out entire exons.
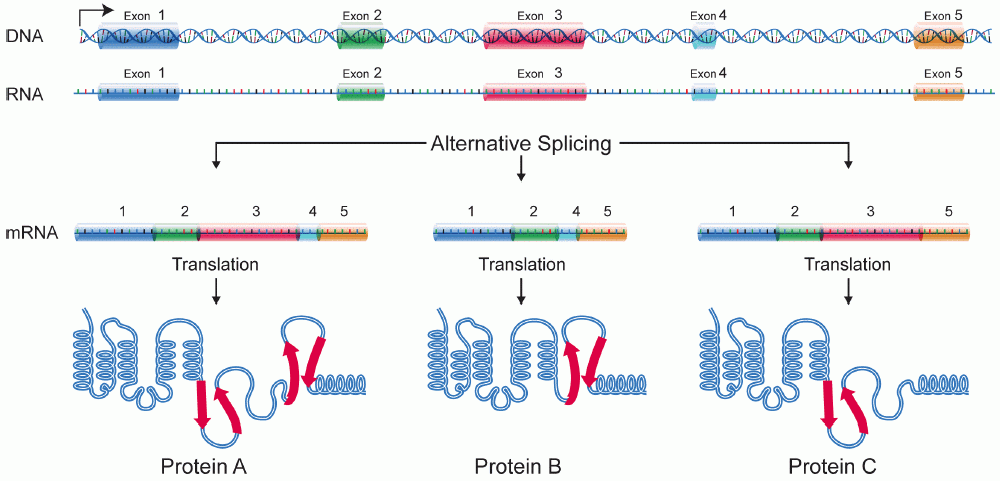
Covalent modifications
The proteins synthesized in translation are subjected to many covalent changes. By these modifications in the amino acids, the proteins may be converted to active form or inactive form. Selected examples of covalent modifications are described below.
Phosphorylation
- It involves the addition of a phosphate (PO4) group to a protein or a small molecule.
- The hydroxyl group containing amino acids of proteins, namely serine, threonine and tyrosine are subjected to phosphorylation.
- The phosphorylation may either increase or decrease the activity of the proteins.
- A group of enzymes called protein kinases catalyse phosphorylation while protein phosphatases are responsible for dephosphorylation (removal of phosphate group).
- Many enzymes that undergo phosphorylation or dephosphorylation are known in metabolisms (e.g. glycogen synthase).
Hydroxylation
- It involves the addition of hydroxyl group to proline of protein.
- During the formation of collagen, the amino acids proline and lysine are respectively converted to hydroxyproline and hydroxylysine. This hydroxylation occurs in the endoplasmic reticulum and requires vitamin C.
Glycosylation
- It involves the addition of saccharide to a protein or a lipid molecule.
- The complex carbohydrate moiety is attached to the amino acids, serine and threonine (O-linked) or to asparagine (N-linked), leading to the synthesis of glycoproteins
Biotinylation
- It involves the addition of biotin to protein or nucleic acid.
Acetylation
- It involves the addition of an acetyl group, usually at the N- terminus of the protein.
Carboxylation
- It involves the addition of carboxyl group to glutamate.
Methylation
- The addition of a methyl group, usually at lysine or arginine residues.
Alkylation
- The addition of an alkyl group (e.g. methyl, ethyl).
Glutamylation
- Covalent linkage of glutamic acid residues to tubulin and some other
Lipoylation
- The attachment of a lipoate functionality Sulfation The addition of a sulfate group to a tyrosine.
Attachment of Carbohydrate Side Chains
- The carbohydrate side chains of glycoproteins are attached covalently during or after synthesis of the polypeptide.
- In some glycoproteins, the carbohydrate side chain is attached enzymatically to Asn residues (N-linked oligosaccharides), in others to Ser or Thr residues (O-linked oligosaccharides).
- Many proteins that function extracellularly, as well as the lubricating proteoglycans that coat mucous membranes, contain oligosaccharide side chains.
Addition of Isoprenyl Groups
- A number of eukaryotic proteins are modified by the addition of groups derived from isoprene (isoprenyl groups).
- A thioether bond is formed between the isoprenyl group and a Cys residue of the protein.
- The isoprenyl groups are derived from pyrophosphorylated intermediates of the cholesterol biosynthetic pathway, such as farnesyl pyrophosphate.
- Proteins modified in this way include the Ras proteins, products of the ros oncogenes and proto-oncogenes, and G proteins, and Iamins, proteins found in the nuclear matrix.
- The isoprenyl group helps to anchor the protein in a membrane.
Addition of Prosthetie Groups
- Many proteins require for their activity covalently bound prosthetic groups.
- Two examples are the biotin molecule of acetylCoA carboxylase and the heme group of hemoglobin or cytochrome c.
Proteolytic Processing
- Many proteins are initially synthesized as large, inactive precursor polypeptides that are proteolytically trimmed to form their smaller, active forms.
- Examples include proinsulin, some viral proteins, and proteases such as chymotrypsinogen and trypsinoge.
Formation of Disulfide Cross-Links
- After folding into their native conformations, some proteins form intrachain or interchain disulfide bridges between Cys residues.
- In eukaryotes, disulfide bonds are common in proteins to be exported from cells.
- The cross-Iinks formed in this way help to protect the native conformation of the protein molecule from denaturation in the extracellular environment, which can differ greatly from intracellular conditions and is generally oxidizing.
Post-translational Processing
Post-translational processing refers to the modifications that occur to proteins after their synthesis is completed. These modifications play a crucial role in shaping the structure, function, and localization of proteins. Here are some key post-translational processing methods:
- Proteolysis: Proteolysis involves the cleavage of specific peptide sequences in a protein. Signal peptidases cleave signal peptides, which are necessary for protein targeting and secretion. Additionally, other polypeptide sequences within the protein may be cleaved to generate the final functional protein. An example of proteolysis is the conversion of inactive insulin into its active form through the removal of a peptide sequence.
- Phosphorylation: Phosphorylation is the addition of phosphate groups to proteins. It is one of the most common post-translational modifications in animal cells and serves as a regulatory mechanism for protein activity. Phosphorylation predominantly occurs on serine, threonine, and tyrosine amino acid residues, and it can activate or deactivate proteins, modulating their functions within the cell.
- Glycosylation: Glycosylation involves the addition of carbohydrate molecules to proteins, forming glycoproteins. Many proteins destined for the plasma membrane or secretion undergo glycosylation. N-glycosylation occurs in the endoplasmic reticulum (ER), where carbohydrate chains attach to asparagine residues, while O-glycosylation occurs in the Golgi complex, where carbohydrate chains attach to serine or threonine residues. Glycosylation can affect protein folding, stability, and interactions with other molecules.
- Sulfation: Sulfation refers to the addition of sulfate molecules to proteins. This modification typically occurs at tyrosine residues and is mediated by enzymes called tyrosylprotein sulfotransferases. Sulfation plays a role in protein-protein interactions, cell adhesion, and modulation of signaling pathways. It is important for the activity of certain proteins, including some secreted factors and extracellular matrix components.
- Methylation: Methylation involves the addition of methyl groups to amino acid side chains within proteins. This modification is mediated by enzymes called methyltransferases and relies on S-adenosyl methionine (SAM) as the methyl group donor. Methylation can regulate protein-protein interactions, protein stability, and gene expression. It commonly occurs on lysine and arginine residues and affects protein function and cellular processes.
- Hydroxylation: Hydroxylation is the addition of a hydroxy group to amino acid side chains. It occurs on specific amino acids, such as proline and lysine, and is catalyzed by hydroxylases. Hydroxylation plays a crucial role in the regulation of various factors, including transcription factors, collagen synthesis, and hypoxia response pathways.
- Other modifications: Other post-translational modifications include SUMOylation, which involves the attachment of small ubiquitin-related modifier (SUMO) proteins to target proteins, affecting their localization and activity. Disulfide bond formation stabilizes protein structure and participates in redox processes. Lipidylation, acetylation, and prenylation are additional modifications that add lipid or acetyl groups to proteins, aiding in their membrane association or protein-protein interactions.
These post-translational processing methods significantly expand the functional diversity of proteins and enable them to perform their roles in various cellular processes.
Significance of Post-transcriptional Modification
- Post-transcriptional modification (PTM) plays a crucial role in the regulation of protein function and the maintenance of cellular homeostasis. While proteins are synthesized through the translation of mRNA by ribosomes, PTMs occur after this process and involve various chemical modifications to the polypeptide chains. These modifications can impact protein activity, localization within the cell, and interactions with other proteins.
- One significant function of PTMs is their involvement in proper protein folding. Calnexin, a type of lectin molecule, binds to glycosylated proteins and assists in their folding. This process ensures that proteins adopt their correct three-dimensional structure, which is essential for their functionality. The addition of glycans to proteins can also confer stability, increasing the protein’s half-life and protecting it from degradation by proteolytic enzymes.
- PTMs are also involved in protein sorting or translocation within the cell. For instance, the presence of phosphorylated mannose residues in a protein directs it to the lysosome, where it can fulfill its specific role. This mechanism ensures that proteins are correctly targeted to their designated cellular compartments, allowing them to carry out their functions effectively.
- Furthermore, PTMs play a crucial role in regulating protein activity and function. Phosphorylation, a reversible PTM, acts as a molecular switch that can activate or deactivate proteins. By adding or removing phosphate groups, the activity of the protein can be modulated, enabling precise control of cellular processes. Another example is acetylation, which regulates a wide range of functions including DNA recognition, protein-protein interactions, and protein stability. These modifications provide a means to fine-tune protein behavior and adapt it to the specific requirements of the cell.
- Redox-dependent PTMs of proteins have emerged as a key signaling system conserved throughout evolution. These modifications, influenced by the cellular redox state, regulate many aspects of cellular homeostasis. Redox modifications can alter protein structure and function, impacting signal transduction pathways, gene expression, and metabolic processes. They play a critical role in cellular responses to oxidative stress and are involved in various diseases and physiological conditions.
- Moreover, PTMs contribute to the diversity and complexity of the proteome. By introducing modifications such as phosphorylation, acetylation, glycosylation, and others, the proteome expands significantly. This increased diversity enables cells to fine-tune protein functions and adapt to a wide range of environmental cues and stimuli.
- Overall, post-transcriptional modifications are of immense significance in cellular processes. They regulate protein folding, stability, activity, and localization. Moreover, PTMs serve as vital components in cell signaling pathways and significantly contribute to the diversity and complexity of the proteome. Understanding the mechanisms and functions of PTMs is essential for unraveling the intricate workings of cellular biology and can have profound implications in various fields, including medicine and biotechnology.
FAQ
What is post-transcriptional modification?
Post-transcriptional modification refers to the chemical modifications that occur to RNA molecules after they have been transcribed from DNA. These modifications can impact RNA stability, localization, and functionality.
What are some common types of post-transcriptional modifications?
Common types of PTMs include alternative splicing, RNA editing, polyadenylation, capping, methylation, and various forms of non-coding RNA modifications.
How does alternative splicing contribute to PTM?
Alternative splicing is a PTM process where different exons within a pre-mRNA can be selectively included or excluded, resulting in the production of multiple protein isoforms from a single gene.
What is the role of RNA editing in post-transcriptional modification?
RNA editing involves the alteration of specific nucleotides within an RNA molecule, leading to changes in the RNA sequence. This modification can impact protein coding potential, RNA structure, and stability.
How does polyadenylation affect RNA molecules?
Polyadenylation involves the addition of a string of adenine nucleotides (poly-A tail) to the 3′ end of an RNA molecule. This modification can influence RNA stability, translation efficiency, and nuclear export.
What is the significance of RNA methylation?
RNA methylation involves the addition of a methyl group to RNA nucleotides. It plays a role in regulating RNA stability, splicing, translation, and interactions with other RNA-binding proteins.
How does capping affect mRNA?
Capping refers to the addition of a modified guanosine nucleotide (m7G cap) to the 5′ end of an mRNA molecule. This modification protects the mRNA from degradation, facilitates mRNA export from the nucleus, and enhances translation efficiency.
What are the functions of non-coding RNA modifications?
Non-coding RNAs, such as transfer RNA (tRNA) and ribosomal RNA (rRNA), undergo various modifications that can impact their structure, stability, and functions in translation and protein synthesis.
Are post-transcriptional modifications reversible?
Yes, some PTMs can be reversible. For example, RNA methylation can be dynamically added or removed, allowing for rapid regulation of RNA function in response to cellular signals or environmental changes.
How do post-transcriptional modifications contribute to gene expression regulation?
PTMs play a crucial role in fine-tuning gene expression. They can affect RNA stability, splicing patterns, translation efficiency, and interactions with regulatory proteins, ultimately influencing the abundance and functionality of proteins in the cell.
References
- Principles of Genetics, D. Peter Snustad, Michael J. Simmons.
- Biochemistry, Dr. U. Satyanarayana, Dr. U. Chakrapani.
- https://en.wikipedia.org/wiki/Post-transcriptional_modification
- https://www.basu.org.in/wp-content/uploads/2021/06/Post-Transcriptional-Modifications_1.pdf
- https://www.slideshare.net/sadiqpa/post-transcriptional-modification
- https://www.slideshare.net/AYSHA007/posttranslational-modifications
- https://www.slideshare.net/saadiaeman/post20translational20modifications
- https://www.slideshare.net/sujay45/posttranslational-modification
- https://plantlet.org/post-transcriptional-modification/
- https://en.wikipedia.org/wiki/Alternative_splicing
