Table of Contents
What is DNA Replication?
In the realm of molecular biology, DNA replication stands as a pivotal biological mechanism responsible for generating two congruent DNA replicas from a singular original DNA molecule. This process is ubiquitous across all living entities, serving as the cornerstone for biological inheritance.
DNA replication is not merely a routine cellular activity; it is indispensable for cellular proliferation during both growth phases and the repair of injured tissues. This ensures that every nascent cell is endowed with its own unique DNA copy, underscoring the inherent capability of cells to undergo division.
The structural foundation of DNA is characterized by a double helix, which is essentially an intertwining of two complementary strands. This double helix configuration is emblematic of a dual-stranded DNA, comprising two linear strands oriented antiparallel to each other, coiling in unison. The intricate process of replication necessitates the separation of these strands.
Subsequently, each strand from the original DNA molecule acts as a template, guiding the synthesis of its complementary strand. This mode of replication, termed as semiconservative replication, ensures that the emergent helix consists of one strand from the original DNA and one newly synthesized strand. The cellular machinery is equipped with meticulous proofreading and error-correction systems, guaranteeing an exceptionally high fidelity in DNA replication.
Within the cellular environment, the onset of DNA replication is demarcated at specific genomic sites, termed as origins of replication. These origins harbor the genome, the repository of an organism’s genetic blueprint. The initiation of replication involves the unwinding of the DNA helix at these origins, facilitated by the enzyme helicase.
This results in the formation of replication forks that extend in both directions from the origin. The replication apparatus comprises a suite of proteins that play instrumental roles in both the initiation and sustenance of DNA synthesis. Of these, DNA polymerase is of paramount importance, orchestrating the synthesis of new strands by appending nucleotides complementary to the template strand. This orchestrated event predominantly transpires during the S-phase of interphase.
Beyond the cellular milieu, DNA replication can also be simulated in vitro. Utilizing DNA polymerases derived from cells and synthetic DNA primers, DNA synthesis can be instigated at predetermined sequences on a template DNA molecule. Techniques such as Polymerase Chain Reaction (PCR), Ligase Chain Reaction (LCR), and Transcription-Mediated Amplification (TMA) exemplify this.
Recent scientific revelations from March 2021 have posited that an archaic form of transfer RNA, integral to the process of translation, might have functioned as a replicator molecule during the nascent stages of life’s evolution.
In summation, DNA replication is an intricate, semiconservative mechanism that transpires during cell division. It employs a parental DNA strand as a template to engender a complementary daughter strand, leveraging a consortium of proteins and enzymes. DNA polymerase plays a central role, catalyzing the formation of the growing DNA chain.
The process is further augmented by other proteins that oversee its initiation, progression, and accuracy. Consequently, DNA replication is a quintessential process, ensuring the preservation of genetic information, facilitating cellular growth, repair, and perpetuating the lineage of organisms.
Definition of DNA Replication
DNA replication is the biological process by which a cell duplicates its DNA molecule, producing two identical copies from one original DNA strand, ensuring the transmission of genetic information during cell division.
Structure of DNA
- The intricate structure of DNA, the molecule responsible for encoding genetic information, was elucidated by James Watson and Francis Crick. Their groundbreaking discovery revealed that DNA adopts a double-helical conformation, characterized by two intertwined strands of nucleic acids. This helical arrangement not only imparts DNA with its distinctive shape but also facilitates its compact organization within cells.
- At the molecular level, DNA is a polymer composed of nucleotides. Each nucleotide unit consists of three fundamental components: a deoxyribose sugar molecule, a phosphate group, and a nucleobase. The nucleobases, which are the information-carrying units of DNA, can be one of four types: Adenine (A), Thymine (T), Guanine (G), or Cytosine (C). These nucleobases engage in specific pairing interactions, with Adenine forming hydrogen bonds with Thymine and Guanine pairing with Cytosine. This base pairing ensures the fidelity of genetic information during processes such as replication.
- The nucleotides are sequentially linked through phosphodiester bonds, establishing a sugar-phosphate backbone that runs the length of each DNA strand. The orientation of this backbone is defined by the 3′ (three-prime) and 5′ (five-prime) ends of the deoxyribose sugar. In the context of the double helix, the two DNA strands are anti-parallel, meaning one strand runs in the 5′ to 3′ direction, while its complementary strand runs in the 3′ to 5′ direction.
- The nucleobases protrude inward from the sugar-phosphate backbone, facilitating their pairing interactions. Adenine and Guanine are classified as purine bases, whereas Cytosine and Thymine are pyrimidines. The hydrogen bonds that mediate the interactions between complementary bases are weaker than the covalent phosphodiester bonds that connect individual nucleotides. This differential in bond strength allows for the temporary separation of the DNA strands during processes like replication.
- Within the cellular nucleus, DNA is meticulously packaged into structures known as chromatins. These structures undergo further condensation to form chromosomes during cell division. Prior to replication, the chromatin fibers undergo a relaxation process, granting the cellular machinery the necessary access to the DNA template.
- In essence, the double-helical structure of DNA, with its anti-parallel strands and specific base pairing, ensures the accurate transmission and expression of genetic information. The robustness of the phosphodiester bonds and the flexibility afforded by the weaker hydrogen bonds between nucleobases underscore the dynamic nature of DNA, allowing it to function as the quintessential molecule of heredity.
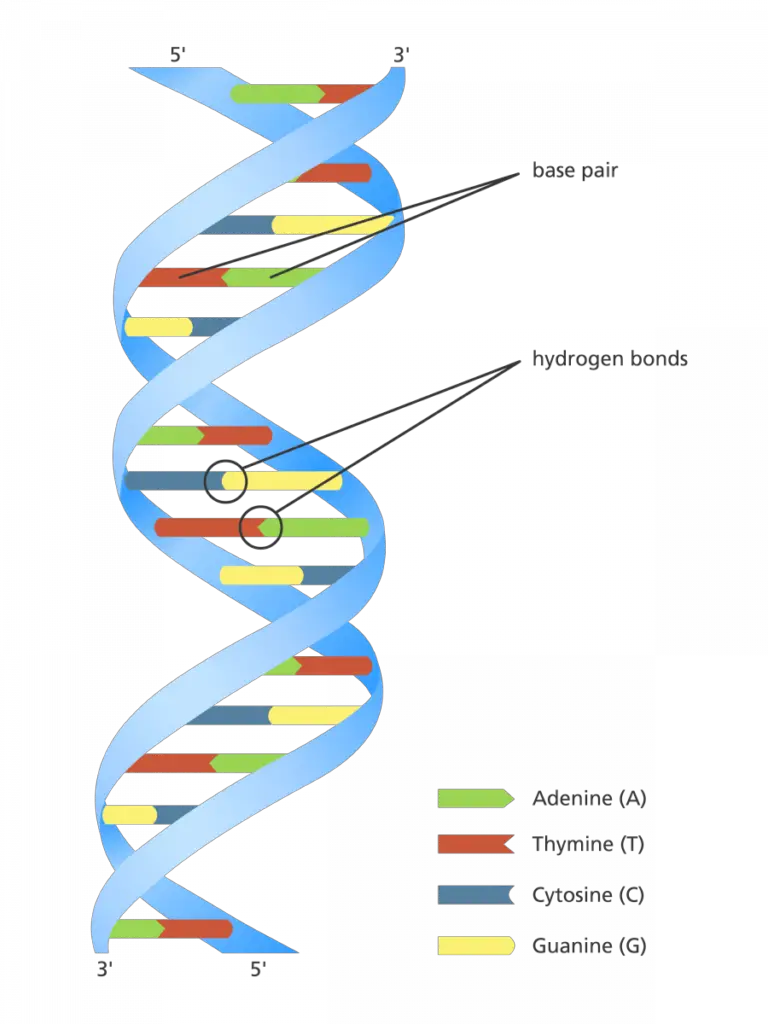
Mechanism of DNA replication/3 steps of DNA replication
DNA replication, a cornerstone of cellular function, is a meticulously orchestrated process ensuring the accurate duplication of the genetic material. This process can be delineated into three cardinal stages:
- Initiation and Strand Separation: The DNA molecule, characterized by its iconic double-helical structure, undergoes unwinding at a designated site termed the ‘origin of replication.’ Here, the intertwined DNA strands are separated, setting the stage for replication.
- Template-Strand Priming: Post strand separation, the individual DNA strands are prepared or ‘primed’ for duplication. This priming process is facilitated by a consortium of enzymes and proteins, which ensure that the strands are rendered amenable to the subsequent synthesis of complementary strands.
- Synthesis and Assembly: The culmination of the replication process is marked by the action of the DNA polymerase enzyme. This enzyme orchestrates the assembly of the nascent DNA strands, ensuring that each nucleotide is accurately paired with its complementary counterpart on the template strand.
While the overarching framework of DNA replication remains consistent across cells, nuances in the process might manifest depending on the specific organism or cell type under consideration. Enzymatic activity is paramount to the fidelity and efficiency of DNA replication. Enzymes catalyze pivotal reactions throughout the replication process, from strand separation to the synthesis of the new DNA strands.
The intricacies of DNA replication have been a focal point of scientific inquiry, given its fundamental role in cellular biology. One of the most extensively studied models in this context is the bacterium Escherichia coli (E. coli). The replication mechanism in E. coli offers valuable insights, as it mirrors, to a significant extent, the replication processes in eukaryotic cells. In E. coli, the initiation of DNA replication is anchored at the oriC locus. The DnaA protein binds to this locus, facilitating the commencement of replication in conjunction with the hydrolysis of ATP.
In summation, DNA replication is a complex yet elegantly coordinated process, underpinned by enzymatic actions and governed by well-defined mechanistic principles. Its understanding not only sheds light on cellular function but also underscores the precision inherent in biological systems.
DNA Replication Enzymes and Proteins
DNA replication is a highly coordinated process, underpinned by a suite of specialized enzymes and proteins that ensure the accurate duplication of the genetic material. Here, we delve into the key enzymes and proteins that play pivotal roles in this intricate process.
- DNA-dependent DNA Polymerase: This enzyme is central to the replication process. It facilitates polymerization, orchestrating the synthesis of the new DNA strand. Deoxyribonucleoside triphosphates serve dual roles as substrates and energy sources. DNA polymerase exists in multiple forms:
- DNA Polymerase I: Involved in DNA repair, it exhibits 5′-3′ polymerase activity, 5′-3′ exonuclease activity, and 3′-5′ exonuclease activity.
- DNA Polymerase II: Responsible for primer extension and proofreading.
- DNA Polymerase III: Catalyzes DNA replication in vivo.
- DNA Helicase (DnaB Protein): This enzyme unwinds the double-stranded DNA, breaking hydrogen bonds and creating replication forks.
- DNA Primase (DnaG Protein): An RNA polymerase variant, it synthesizes RNA primers, which are essential for initiating DNA replication.
- Topoisomerase/DNA Gyrase: These enzymes alleviate the supercoiling and tangling of DNA by introducing and subsequently resealing breaks in the DNA strands.
- Exonucleases: These enzymes excise nucleotides from the ends of DNA strands, ensuring fidelity in replication.
- DNA Ligase: It seals gaps between nucleotides, forming phosphodiester bonds and ensuring the continuity of the DNA strand.
- Single-stranded Binding Proteins: By binding to single-stranded DNA, these proteins prevent the formation of secondary structures, ensuring the DNA remains accessible for replication.
- Telomerase: In eukaryotic cells, this enzyme stabilizes chromosomes by adding specific DNA sequences to chromosome ends after each division.
- Tus Protein: This protein plays a role in the terminal stages of replication.
- SSB Protein: Post helicase action, this protein binds to the single-stranded DNA, ensuring its stability.
- DnaA Protein: Essential for the initiation of replication, this protein binds to the replication origin.
- Helicase Loader (Dna C Protein): In conjunction with DnaA, this protein guides helicase to the DNA template.
| Enzyme Name | Function |
|---|---|
| DNA Polymerase | Synthesizes new DNA strands by adding nucleotides to the growing DNA chain. |
| DNA Helicase | Unwinds the double-helical structure of DNA, allowing DNA replication to commence. |
| DNA Primase | Synthesizes short RNA primers, which act as templates for the initiation of DNA replication. |
| DNA Ligase | Joins DNA fragments together by forming phosphodiester bonds between nucleotides. |
| Exonuclease | Removes nucleotide bases from the end of a DNA chain and proofreads the new strands. |
| Topoisomerase (or DNA Gyrase) | Solves topological stress caused during unwinding by cutting one or both DNA strands. |
| Telomerase | Adds specific DNA sequence to the telomeres of chromosomes after they divide. |
DNA Replication Steps (DNA Replication Process)
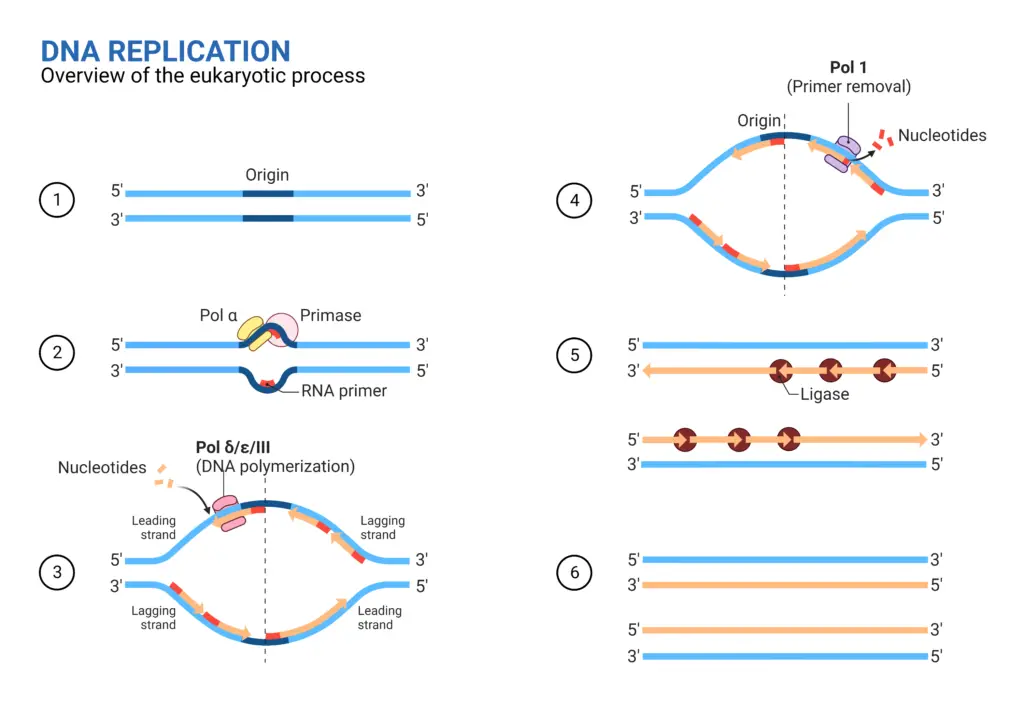
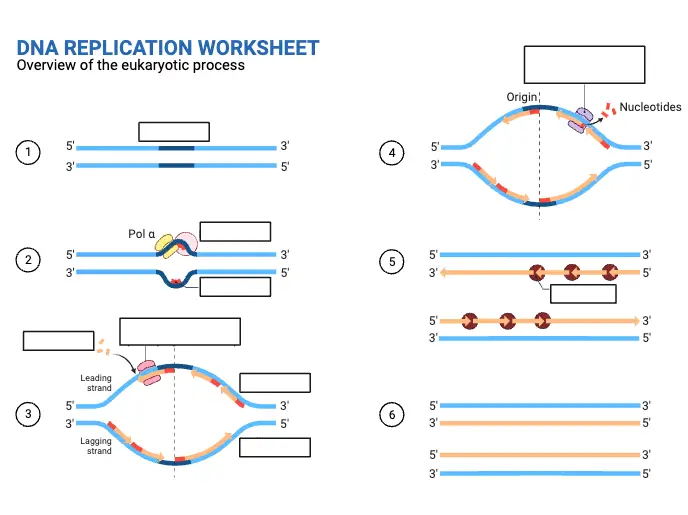
1. Initiation
The initiation of DNA replication marks the commencement of the intricate process of duplicating the genetic material. This pivotal stage is characterized by a series of coordinated events that ensure the accurate and efficient replication of DNA.
- Origins of Replication: DNA replication is instigated at specific loci on the template strand termed as ‘origins.’ These regions are genetically coded to serve as the starting points for the replication process.
- Role of Initiator Proteins: The origins are recognized and bound by initiator proteins. These proteins play a crucial role in recruiting a suite of additional proteins essential for replication, collectively forming a replication complex around the DNA origin.
- Formation of Replication Forks: Multiple sites, known as replication forks, are established where DNA replication is initiated. These forks represent the active zones of DNA unwinding and synthesis.
- Action of DNA Helicase: Central to the replication complex is the DNA helicase enzyme. Its primary function is to unwind the iconic double-helical structure of DNA, thereby exposing the two template strands for replication. The mechanism by which DNA helicase operates involves the hydrolysis of ATP. This energy-driven process facilitates the breaking of hydrogen bonds between nucleobases, leading to the separation of the DNA strands.
- Synthesis of RNA Primers: Concurrently, the DNA primase enzyme synthesizes short RNA primers. These primers are essential as they provide the starting point for the DNA polymerase enzyme to begin its synthesis activity.
- Role of DNA Polymerase: DNA polymerase is the workhorse of the replication process. Once activated by the RNA primers, this enzyme commences the synthesis of the new DNA strand, ensuring that each nucleotide is accurately paired with its complementary counterpart on the template strand.
In essence, the initiation phase of DNA replication is a meticulously coordinated event, setting the stage for the subsequent phases of elongation and termination. The interplay of various enzymes and proteins ensures that the genetic blueprint is accurately and efficiently duplicated, underscoring the precision inherent in cellular processes.
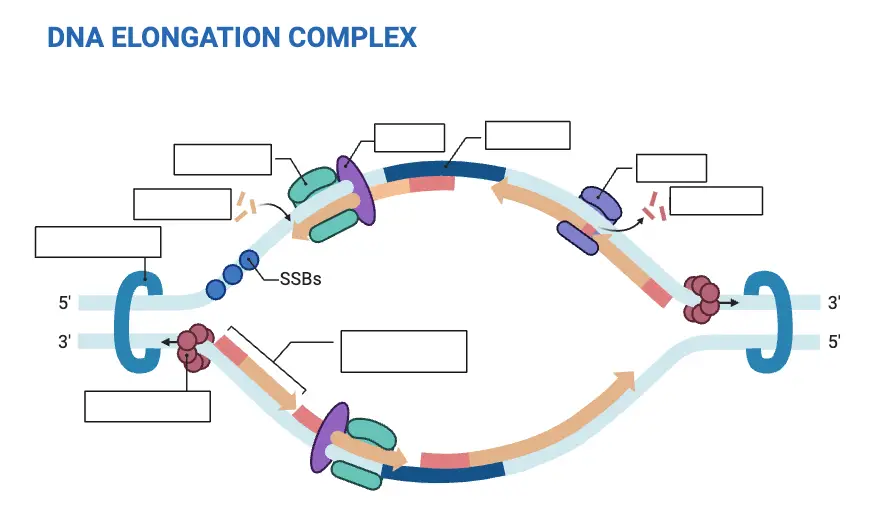
2. Elongation
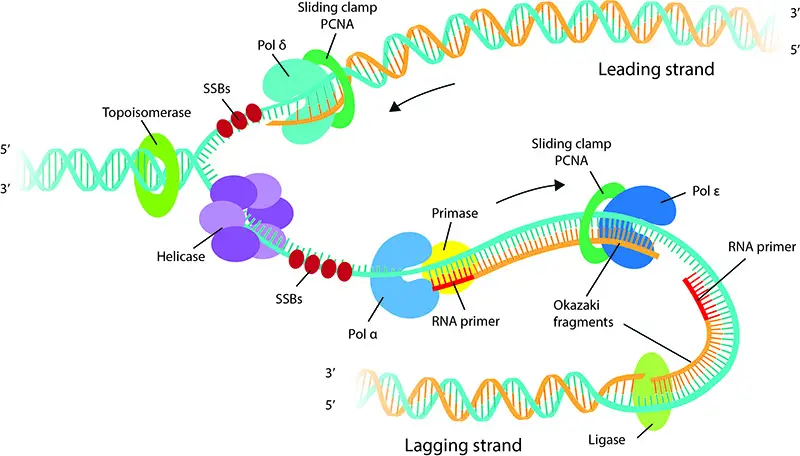
Elongation is a pivotal phase in DNA replication, characterized by the synthesis of the new DNA strand. This process is facilitated by a suite of enzymes and proteins that work in concert to ensure the accurate and efficient duplication of the genetic material.
- DNA Polymerase Activity: At the heart of the elongation process is the DNA polymerase enzyme. It latches onto the unzipped template strand, using the short RNA primer as a starting point. By harnessing free nucleotides, DNA polymerase extends the primer, synthesizing a new strand complementary to the template. This synthesis occurs by adding nucleotides to the 3′ end of the growing strand.
- Leading Strand Synthesis: One of the template strands is read in the 3′ to 5′ direction. Consequently, DNA polymerase synthesizes the complementary strand continuously in the 5′ to 3′ direction. This continuously synthesized strand is termed the ‘leading strand.’
- Lagging Strand Synthesis: The synthesis of the complementary strand for the other template (oriented 5′ to 3′) is more complex. DNA primase periodically synthesizes short RNA primers along this template. DNA polymerase then extends these primers, creating short DNA segments known as ‘Okazaki fragments.’ Given its discontinuous nature, this strand is termed the ‘lagging strand.’
- Okazaki Fragments: These are short, discontinuous fragments synthesized on the lagging strand. Their formation is a consequence of the antiparallel nature of DNA and the directionality of DNA polymerase activity.
- Primer Replacement: The RNA nucleotides of the short RNA primers are transient and must be replaced with DNA nucleotides to ensure the integrity of the newly synthesized strand.
- Role of DNA Ligase: Once the RNA primers are replaced with DNA nucleotides, gaps remain between the Okazaki fragments. DNA ligase functions to seal these gaps, creating a continuous DNA strand.
In summary, the elongation phase of DNA replication is a testament to the precision and coordination inherent in cellular processes. The interplay of enzymes ensures that the genetic blueprint is accurately duplicated, with each nucleotide meticulously added to ensure the fidelity of the replication process.
4. Termination
The termination phase of DNA replication signifies the culmination of the intricate process of duplicating the genetic material. This phase ensures the fidelity and integrity of the newly synthesized DNA strands. Here’s a detailed breakdown of the events that transpire during this phase:
- Role of Exonuclease: Post the synthesis of both continuous and discontinuous strands, an enzyme called exonuclease comes into play. Its primary function is to meticulously remove all RNA primers that were initially laid down on the original strands.
- Primer Replacement: Once the RNA primers are excised, the vacant sites are promptly filled with the appropriate nucleotide bases, ensuring the continuity of the DNA strand.
- Proofreading Mechanism: Another variant of exonuclease undertakes the critical task of proofreading the newly synthesized strands. It scans for any anomalies or errors that might have occurred during the synthesis phase. Upon detection, it excises the incorrect nucleotide and facilitates its replacement with the correct one.
- Okazaki Fragment Ligation: The discontinuous fragments synthesized on the lagging strand, known as Okazaki fragments, need to be cohesively joined to form a continuous strand. The enzyme DNA ligase is instrumental in this process, sealing the gaps between these fragments.
- Telomere Synthesis: The extremities of the parent DNA strand are adorned with repetitive DNA sequences termed telomeres. These sequences serve a protective function, preventing chromosomes from undergoing undesirable fusions. The enzyme responsible for synthesizing these telomeres is telomerase. It elongates the ends of the chromosomes by adding telomere sequences.
- Formation of the Double Helix: As the termination phase reaches its conclusion, the parent strand and its complementary counterpart wind together, adopting the iconic double helical structure. This results in the formation of two DNA molecules, each comprising one strand from the original molecule and one newly synthesized strand.
In essence, the termination phase of DNA replication underscores the precision and meticulousness inherent in cellular processes. It ensures that the genetic information is not only accurately duplicated but also safeguarded for subsequent cellular activities.
| Step Name | Description |
|---|---|
| 1. Initiation | DNA synthesis begins at designated sites in the template strand’s coding regions called origins. Initiator proteins target these sites and assemble a replication complex. |
| 2. Elongation | DNA polymerase grows the new DNA daughter strand by attaching to the original unzipped template strand and the initiating short RNA primer. One strand is synthesized continuously (leading strand) while the other is synthesized in fragments (lagging strand). |
| 3. Termination | After synthesis, exonuclease removes all RNA primers from the original strands. The primers are replaced with the right nucleotide bases. DNA ligase then joins the Okazaki fragments. |
What are Okazaki fragments?
- In the realm of molecular biology, the synthesis of new DNA strands is a meticulous process, given the antiparallel nature of DNA. This antiparallel configuration means that while one strand, known as the leading strand, is synthesized continuously in the 5′ to 3′ direction, its counterpart, the lagging strand, faces a more intricate synthesis process.
- DNA polymerase, the enzyme responsible for catalyzing the polymerization of deoxyribonucleotide triphosphates (dNTPs), operates exclusively in the 5′ to 3′ direction. This directional limitation poses a challenge for the continuous synthesis of the lagging strand.
- To circumvent this, the lagging strand is synthesized in a series of short, discontinuous segments, termed Okazaki fragments. These fragments are synthesized in a direction opposite to the replication fork’s movement.
- The genesis of each Okazaki fragment is marked by an RNA primer, a short RNA sequence that provides the starting point for DNA synthesis. This primer is synthesized de novo by an enzyme called primase, which crafts short RNA sequences, typically 3-10 nucleotides in length, that are complementary to the lagging strand template at the replication fork.
- Following the primer’s establishment, DNA polymerase extends these RNA primers, leading to the formation of the Okazaki fragments.
- A notable feature of the freshly synthesized lagging strand is the presence of RNA-DNA joints, underscoring RNA’s pivotal role in DNA replication. To transform these fragments into a continuous DNA strand, the RNA primers are subsequently removed and replaced with DNA. The final step in this intricate dance involves the enzyme DNA ligase, which seamlessly joins the Okazaki fragments, resulting in a unified, continuous lagging strand.
What is Replication Fork?
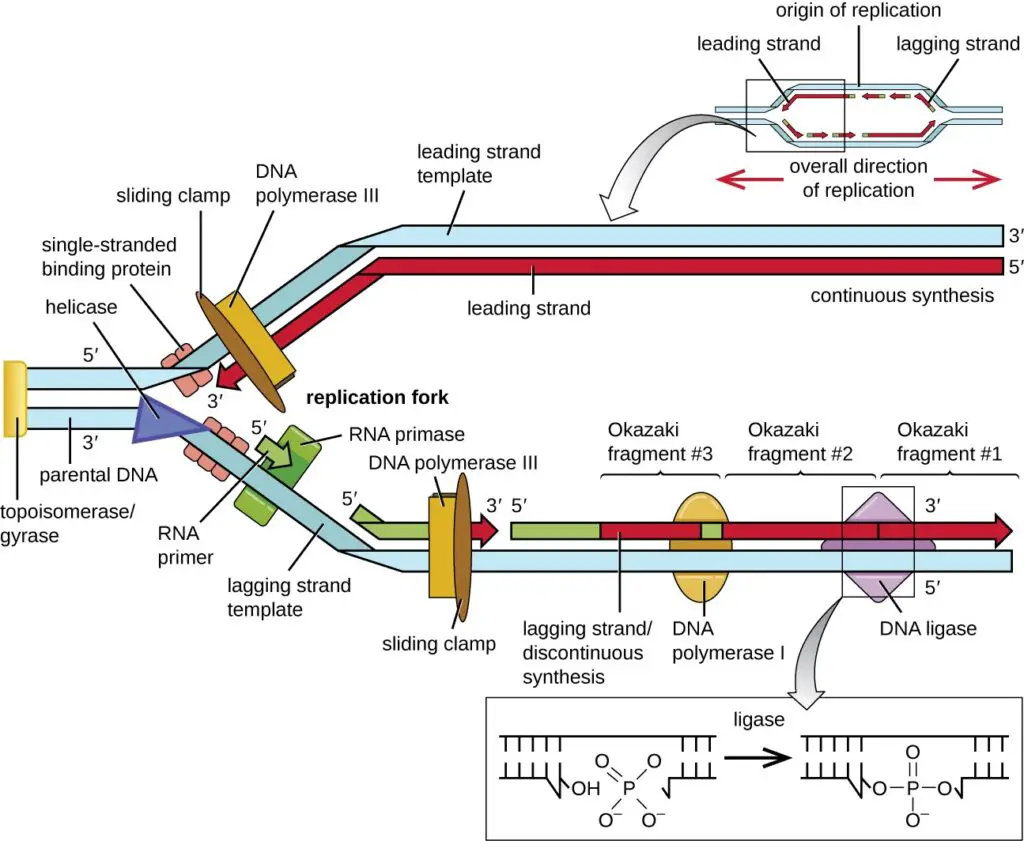
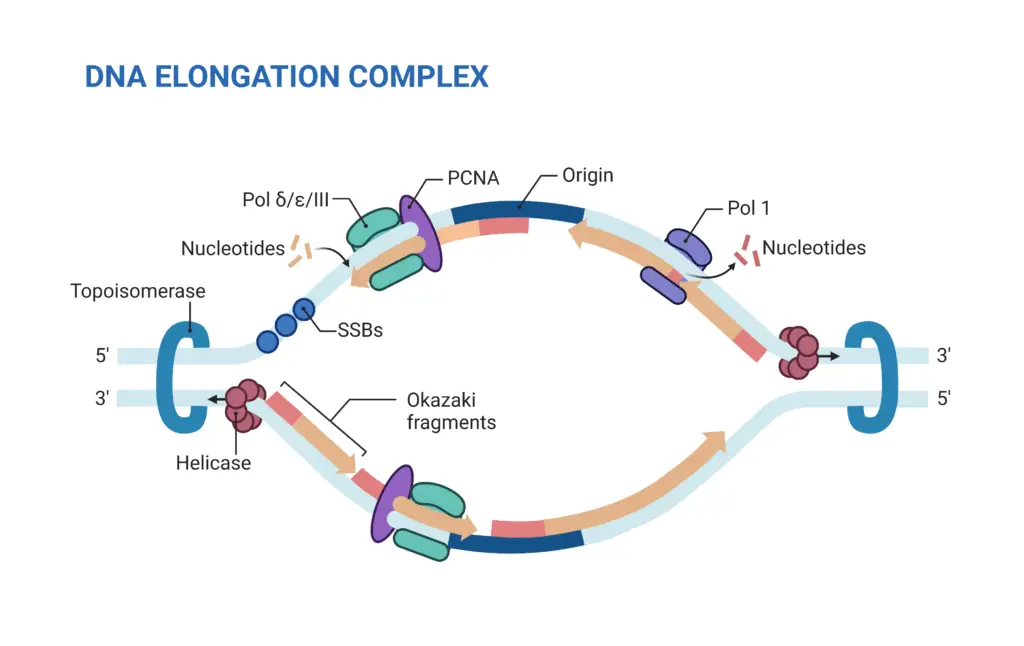
The replication fork is a critical structure that emerges during the DNA replication process. It is formed when specific enzymes, known as helicases, act on the DNA double helix, disrupting the hydrogen bonds that hold the two DNA strands together. This enzymatic action results in a “Y”-shaped structure, with each arm of the “Y” representing a single-stranded DNA template. These templates are pivotal for the synthesis of the leading and lagging strands.
1. Leading Strand: The leading strand is synthesized continuously in the direction of the advancing replication fork. DNA synthesis on this strand progresses in the 5′ to 3′ direction, which aligns with the movement of the replication fork.
2. Lagging Strand: In contrast, the lagging strand faces a more complex synthesis process. Its orientation is opposite to the replication fork’s movement, leading to its synthesis in a discontinuous manner. This strand is constructed in short segments known as Okazaki fragments. The synthesis begins with a primase enzyme generating a short RNA primer. Following this, DNA polymerase extends these primed segments. Subsequently, RNA primers are replaced with DNA, and the DNA fragments are linked together by the enzyme DNA ligase.
Dynamics at the Replication Fork:
- Helicase Activity: The helicase, a hexameric protein complex, encircles one of the DNA strands. In eukaryotes, it surrounds the leading strand, while in prokaryotes, it encompasses the lagging strand.
- Topological Challenges: As the helicase unwinds the DNA, it induces rotational tension in the DNA ahead of the fork. To counteract this torsional strain, topoisomerases temporarily cleave the DNA strands, introducing negative supercoils that alleviate the tension.
- Single-strand Binding Proteins: These proteins bind to the exposed single-stranded DNA, preventing it from forming secondary structures that could hinder DNA polymerase activity.
- Histone Dynamics: DNA in eukaryotes is wrapped around histone proteins. During replication, histone chaperones disassemble and reassemble these histone-DNA complexes to ensure the newly synthesized DNA retains its chromatin structure.
- Clamp Proteins: These proteins function as sliding clamps, enhancing the processivity of DNA polymerase. They ensure the polymerase remains attached to the template. When the polymerase reaches the template’s end or encounters double-stranded DNA, the clamp undergoes a conformational shift, releasing the polymerase.
In summary, the replication fork is a dynamic and complex structure essential for the accurate and efficient replication of DNA. It involves a coordinated interplay of various enzymes and proteins to ensure the faithful duplication of the genetic material.
DNA Replication Process in Prokaryotes
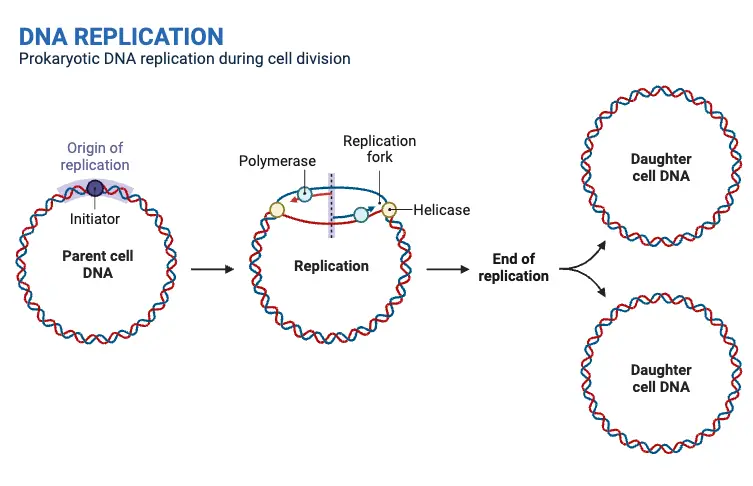
In prokaryotes DNA replication happens to be a simple process when compared to the eukaryotes. Here’s an outline about the replication of DNA in prokaryotes.
- Initiation: The process of DNA replication starts at a specific location known as the site of origin for replication (oriC). The site is identified by initiator proteins, which connect to it and initiate the unwinding process of the double helix of DNA.
- Unwinding: After the initiator proteins are bound to the point of origin of DNA, they trigger DNA strands unwind and form a bubble of replication. Unwinding is made easier by helicases or enzymes that separate the two strands of DNA molecules.
- Elongation: Once the DNA strands not wound, DNA polymerase, an enzyme, begins the creation and creation of DNA strands. DNA polymerase is a nucleotide-binding enzyme that adds nu the growing DNA strands adhering to the base-pairing rule (A with T as well as G and C) and using already existing DNA strands for templates. One strand, referred to as”the lead strand,” is made continuously from 5′-3 direction. The other strand, known as the leading strand, is produced intermittently in small pieces called Okazaki fragments.
- Priming: Prior to the time that DNA polymerase is able to initiate replication the RNA primer is made via an enzyme known as primase. The RNA primer is an initial point of entry that allows DNA polymerase to start adding nucleotides.
- Synthesis and proofreading: DNA polymerase is a additional nucleotides to the growing DNA strands, which extends them from 5′-3 direction. When it synthesizes new DNA strands also functions as a proofreader that assists in identifying and correct any mistakes that could be made.
- Termination: DNA replication continues bidirectionally up to the point that replication forks of both directions meet. After that the termination proteins play a role in stopping replication and making sure that the whole DNA molecule has been duplicated.
In the end, DNA replication within prokaryotes is an extremely fast and efficient process because of the small size of the genome. The ease that the DNA replication in prokaryotes enables rapid replication and the rapid growth of prokaryotic cell.
DNA Replication Process in Eukaryotes
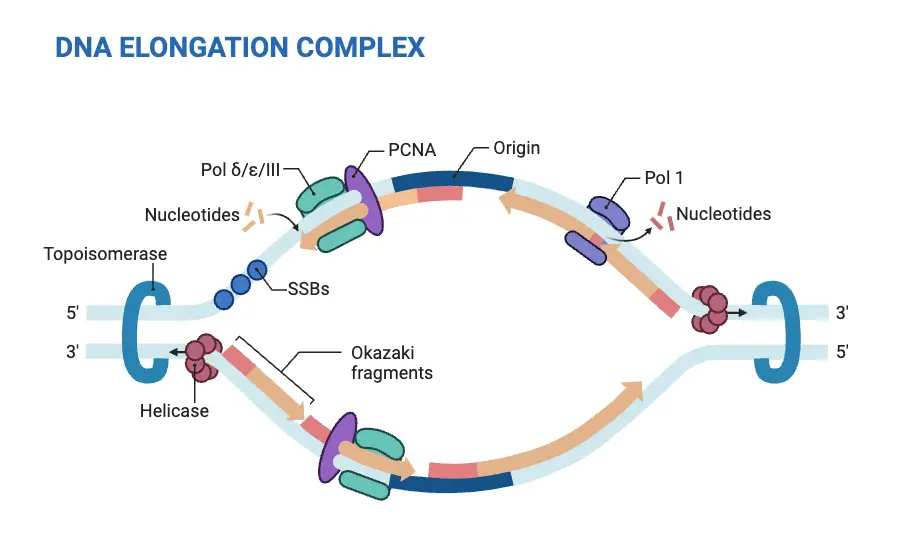
In eukaryotes DNA replication is an intricate and highly controlled process that happens in the nucleus cell. Here’s a brief outline of DNA replication that occurs in the eukaryotes
- Initialization: DNA replication starts at multiple locations, referred to as the sources of replication scattered throughout the genome. The process of starting replication can be controlled through a set of proteins referred to as Pre-Replication Complex (pre-RC) that recognizes and binds with the origins.
- Unwinding: When the pre-RC has formed at the point of origin for replication, the helicase enzymes become activated and unwind and break the DNA double Helix. This results in two replication forks. the DNA strands of parents are separated and act as DNA synthesis templates.
- Elongation: DNA synthesizing occurs at every replication fork, in a bidirectional fashion. Primase creates short DNA primers, which serve as the foundation for DNA polymerases that can begin making the new DNA-strands. DNA polymerases are able to add complementary nucleotides to the growing DNA strands adhering to the base-pairing rule (A with T as well as G and C) and then extend the strands along the 5′-3 direction. One strand, referred to as”the lead strand,” is created constantly in the direction of the replication fork and the second strand, also known as the lagging one, is created in irregularly-spaced Okazaki fragments.
- Replication Complexes: When DNA replication is progressing the large protein complex known as the replisome is assembled at every replication fork. The replisome is comprised of DNA polymerases and helicases, primases and other proteins that help coordinate and speed up the process of replication.
- Repair and Proofreading: DNA polymerases possess the ability to proofread, which permits them to identify and fix any errors that might be made during DNA synthesizing. In addition DNA repair mechanisms work throughout replication and afterwards to repair any errors or damage to DNA that could occur.
- Telomeres: Eukaryotic DNA has specialized sequences known as telomeres that are located at the end of the chromosomes. Telomeres are involved in ensuring the integrity of ends of the chromosomes in the course of replication.
- Termination: The DNA replication process is an ongoing process that takes place during all of the S stage of cell development. It ends when replication forks of adjacent origins join to complete replication process of the entire the chromosome.
Replication of DNA in the eukaryotes can be more complex and requires a variety of protein complexes and enzymes, in comparison to prokaryotes. The coordination and control of these components assures exact as well as complete reproduction of genome of eukaryotes which allows for the reliable transfer of information from genetics to the daughter cells.
Enzymes involved in DNA Replication in the prokaryote, E. coli
| E. coli Gene | Enzyme/Protein Function | Description |
|---|---|---|
| dnaA | Initiator Protein | Melts DNA at oriC, exposing two template ssDNA strands |
| dnaB | Helicase | Unwinds the DNA helix at the front end of each replication fork during replication |
| dnaC | Helicase Loader | Loads the DnaB Helicase onto the ssDNA template strands |
| dnaG | Primase | Synthesizes RNA primers used to initiate DNA synthesis |
| dnaE | α-Catalytic Subunit of DNA Polymerase III | Catalytic subunit of the main replicative polymerase during DNA replication |
| dnaQ | ε-Editing Subunit of DNA Polymerase III | Editing subunit of the main replicative polymerase during DNA replication |
| dnaN | β-clamp subunit of DNA Polymerase III | Clamping subunit of the main replicative polymerase during DNA replication |
| polA | DNA Polymerase I | Processes Okazaki fragments and also fills in gaps during DNA repair processes |
| polB | DNA Polymerase II | Proofreading and editing, especially on lagging strand synthesis and some involvement in DNA repair |
| ssb | Single Stranded Binding Proteins (SSB) | Bind with single-stranded regions of DNA in the replication fork and prevent the strands from rejoining |
| gyrA, gyrB | DNA Gyrase | Type II Topoisomerase involved in relieving positive supercoiling tension caused by the action of Helicase |
| parC, parE | Topoisomerase IV | Type II Topoisomerase involved in decatenation of daughter chromosomes during DNA replication |
| ligA | DNA Ligase | Fixes nicks in the DNA backbone during DNA replication, DNA damage, and DNA repair processes |
Differences between DNA replication in Eukaryotes and Prokaryotes
DNA replication is a fundamental process in all living organisms, ensuring that genetic information is passed from one generation to the next. Both prokaryotic and eukaryotic cells follow a semi-conservative method of DNA replication, but the process varies in complexity due to differences in their size and genome structure.
Similarities:
- Unwinding of DNA: Both prokaryotes and eukaryotes utilize specific enzymes to unwind the DNA double helix in preparation for replication.
- Role of DNA Polymerase: In both types of organisms, the enzyme DNA polymerase plays a pivotal role in synthesizing the new DNA strands.
- Semi-conservative Replication: Both prokaryotes and eukaryotes follow a semi-conservative replication model, where each of the two resulting DNA molecules has one old and one new strand.
- Okazaki Fragments: The lagging strand in both organisms is synthesized discontinuously in the form of Okazaki fragments.
- RNA Primers: Initiation of DNA replication in both prokaryotes and eukaryotes requires short RNA primers.
Differences:
- Genome Size: Eukaryotes typically have a much larger genome compared to prokaryotes. Eukaryotic cells possess about 25 times more DNA than prokaryotic cells.
- Origins of Replication: Eukaryotic DNA replication begins at multiple origins of replication, and it occurs unidirectionally within the cell’s nucleus. In contrast, prokaryotic DNA has a single origin of replication, and replication proceeds bidirectionally.
- DNA Polymerases: Eukaryotic cells have multiple types of DNA polymerases, whereas prokaryotes generally have fewer types.
- Rate of Replication: DNA replication is slower in eukaryotes, taking up to 400 hours, while in prokaryotes, it’s much faster, completing in about 40 minutes.
- Telomere Replication: Eukaryotic chromosomes have linear structures with telomeres at their ends, requiring a special mechanism for replicating telomeres. Prokaryotes, with their circular chromosomes, do not have telomeres.
- Timing of Replication: Eukaryotic cells replicate their DNA only during the S-phase of the cell cycle. In contrast, prokaryotic cells, not being bound by a defined cell cycle, can replicate their DNA almost continuously.
In conclusion, while there are foundational similarities in the DNA replication process of prokaryotes and eukaryotes, the differences arise due to the complexities of eukaryotic cellular and genomic structures.
| Aspect | Eukaryotes | Prokaryotes |
|---|---|---|
| Unwinding of DNA | Both utilize enzymes to unwind DNA. | Both utilize enzymes to unwind DNA. |
| Role of DNA Polymerase | DNA polymerase synthesizes new strands. | DNA polymerase synthesizes new strands. |
| Replication Model | Semi-conservative replication. | Semi-conservative replication. |
| Okazaki Fragments | Present in the lagging strand. | Present in the lagging strand. |
| RNA Primers | Required for initiation. | Required for initiation. |
| Genome Size | Larger genome (about 25 times more DNA). | Smaller genome. |
| Origins of Replication | Multiple origins, unidirectional. | Single origin, bidirectional. |
| DNA Polymerases | Multiple types. | One or two types. |
| Rate of Replication | Slower (up to 400 hours). | Faster (about 40 minutes). |
| Telomere Replication | Special mechanism for telomeres. | No telomeres (circular chromosomes). |
| Timing of Replication | During S-phase of the cell cycle. | Almost continuously. |
Functions of DNA
DNA, or deoxyribonucleic acid, is the hereditary material in humans and almost all other organisms. It carries the instructions necessary for the development, functioning, growth, and reproduction of all known living organisms and many viruses. Here are the primary functions of DNA:
- Genetic Blueprint: DNA serves as a blueprint for the entire organism, determining traits ranging from eye color to susceptibility to certain diseases.
- Protein Synthesis: DNA contains the instructions needed to construct and maintain the organism. These instructions are used to build proteins, which are complex molecules that do much of the work in the body.
- Replication: DNA can make copies of itself, ensuring that genetic information can be passed from cell to cell during growth and to new generations during reproduction.
- Mutation Storage: While often considered in a negative context, mutations are a source of genetic variation, which can be beneficial. They are essential for evolution, as they provide the raw material for natural selection.
- Regulation: DNA contains regions (genes) that are used to produce RNA molecules without being used to make proteins. Some of these RNA molecules play roles in the regulation of gene expression, ensuring that genes are turned on and off at the right times.
- Storage of Information: DNA is a stable molecule that stores genetic information for the lifetime of an organism and can be passed from one generation to the next.
- Repair and Recombination: DNA has mechanisms to repair any damage that occurs to its structure. It can also recombine, allowing for genetic diversity in offspring.
- Evolution: Over time, changes (mutations) in DNA accumulate, leading to the evolution of species.
- Cellular Processes Regulation: DNA, through the process of transcription and translation, regulates various cellular processes, ensuring the cell functions correctly.
- Chromosome Structure: DNA, along with proteins, makes up the structure of chromosomes, which are found in the nucleus of eukaryotic cells.
Importance of DNA replication
DNA replication is a fundamental biological process that ensures the transmission of genetic information from one generation to the next. This intricate mechanism is responsible for the accurate duplication of the genetic code, which is stored in the form of DNA molecules.
- Cellular Functionality: DNA contains the blueprints for synthesizing proteins and enzymes, which are pivotal for cellular operations. Without accurate DNA replication, cells might lack the essential machinery to produce these vital molecules.
- Cell Division: As cells divide, it is imperative that each daughter cell receives an accurate copy of the DNA. DNA replication ensures that every new cell has the same genetic information as its predecessor.
- Maintaining Genetic Integrity: Faulty DNA replication can lead to serious complications. If a daughter cell is devoid of the correct genetic information, it may not function properly, leading to potential health issues in multicellular organisms.
- Preservation of Diploid State: In autosomal cells, DNA replication is crucial for maintaining the diploid state, ensuring that each cell has two sets of chromosomes.
- Genetic Transmission and Evolution: DNA replication is the mechanism by which genetic information is passed from one generation to another. While the process is highly accurate, occasional errors, or mutations, can occur. These mutations, though often subtle, are the driving force behind evolution and the vast biological diversity observed in nature.
- Individual Uniqueness: While DNA replication ensures species continuity by producing nearly identical copies of DNA, the minor variations that do occur contribute to the uniqueness of each individual organism.
- Replication Mechanisms: Among the various potential mechanisms of DNA replication, the semi-conservative mode is the most widely accepted. In this method, each of the two parent DNA strands serves as a template for the synthesis of a new complementary strand, ensuring the fidelity of genetic transmission.
What is DNA replication stress?
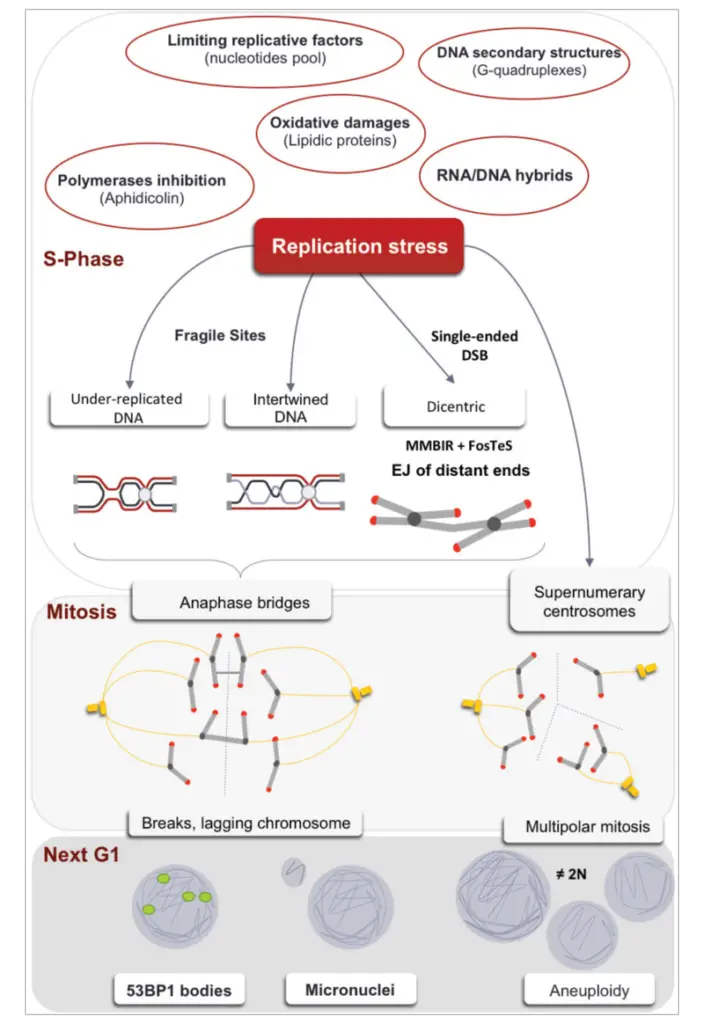
DNA replication stress refers to the challenges and disruptions that arise during the DNA replication process, potentially leading to stalled replication and the formation of halted replication forks. This stress can be attributed to various factors and events, each posing a unique challenge to the smooth progression of replication.
- Complex DNA Structures: Certain regions of the DNA may have unusual structures that can impede the replication machinery, causing it to stall or slow down.
- Mismatched Ribonucleotides: The incorporation of mismatched ribonucleotides can disrupt the replication process, leading to potential errors in the newly synthesized DNA strand.
- Concurrent Processes: Tensions can arise when replication and transcription, two major DNA-dependent processes, occur simultaneously on the same DNA template. This can lead to conflicts and disruptions in both processes.
- Limited Replication Factors: The scarcity of essential replication factors can hinder the replication process, causing it to pause or stall.
- Fragile DNA Sites: Certain regions in the DNA, known as fragile sites, are more susceptible to breakage and can pose challenges during replication.
- Oncogene Activity: Overexpression or continuous activation of oncogenes can induce replication stress, contributing to genomic instability.
- Chromatin Accessibility: In some regions, the DNA may be tightly packed within chromatin, making it less accessible to the replication machinery.
- Role of Regulatory Proteins: Proteins like ATM (ATM serine/threonine kinase) and ATR play a crucial role in mitigating replication stress. They are activated in response to DNA damage and help stabilize the replication process.
When replication forks stall and are not promptly stabilized by regulatory proteins, they can collapse. This collapse triggers repair mechanisms that aim to restore the replication fork and rectify any damage to the DNA ends. In essence, DNA replication stress underscores the delicate balance and precision required in DNA replication, emphasizing the importance of regulatory mechanisms in maintaining genomic integrity.
Why is DNA called a Polynucleotide Molecule?
DNA, or deoxyribonucleic acid, is termed a polynucleotide molecule due to its structural composition. At its core, DNA is composed of smaller units known as nucleotides. Each nucleotide consists of three components: a phosphate group, a deoxyribose sugar, and one of four nitrogenous bases – deoxyadenylate (A), deoxyguanylate (G), deoxycytidylate (C), or deoxythymidylate (T).
When numerous nucleotides link together through phosphodiester bonds, they form a long chain. This chain, due to its composition of multiple nucleotides, is referred to as a polynucleotide. Given that DNA possesses two such chains that coil around each other to form the iconic double helix structure, it is aptly described as a molecule comprising two polynucleotide chains. This intricate arrangement ensures the storage and transmission of genetic information within organisms.
What is Chargaff’s Rule?
Erwin Chargaff, a prominent biochemist, made a pivotal observation regarding the composition of DNA, leading to what is now known as Chargaff’s Rule. Through his research, Chargaff discerned a consistent relationship between the nitrogenous bases present in DNA. Specifically, he noted that the quantity of adenine (A) always equaled the quantity of thymine (T), and similarly, the quantity of cytosine (C) was always equivalent to the quantity of guanine (G).
Mathematically, this relationship can be represented as: A = T and C = G
This discovery was foundational in understanding the pairing mechanism of these bases in the DNA double helix structure. Chargaff’s Rule essentially posits that in any given DNA molecule, the proportion of purine bases (A and G) will always match the proportion of pyrimidine bases (T and C), maintaining the structural integrity and consistency of the DNA across diverse organisms.
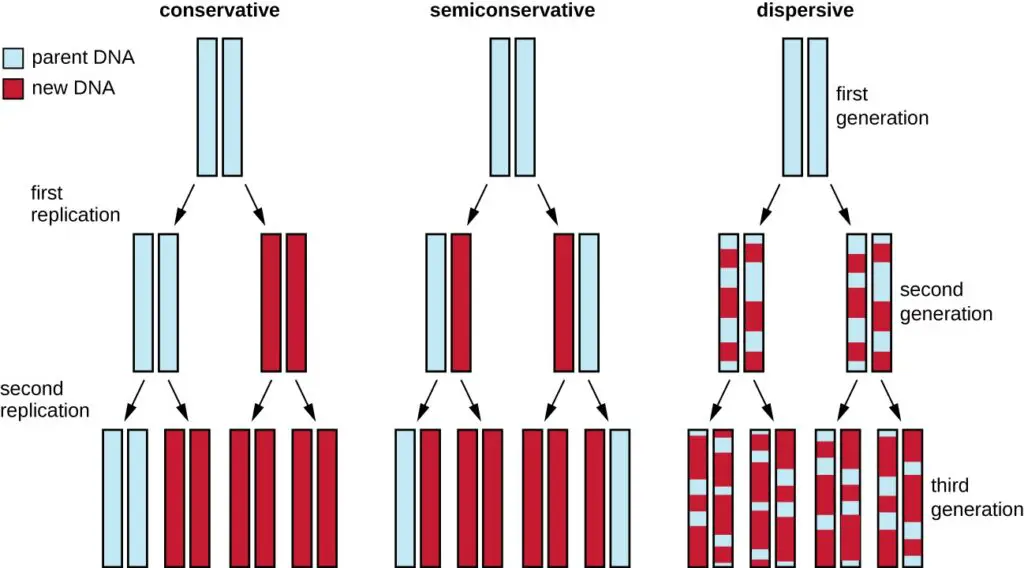
Why dna replication is called semiconservative?
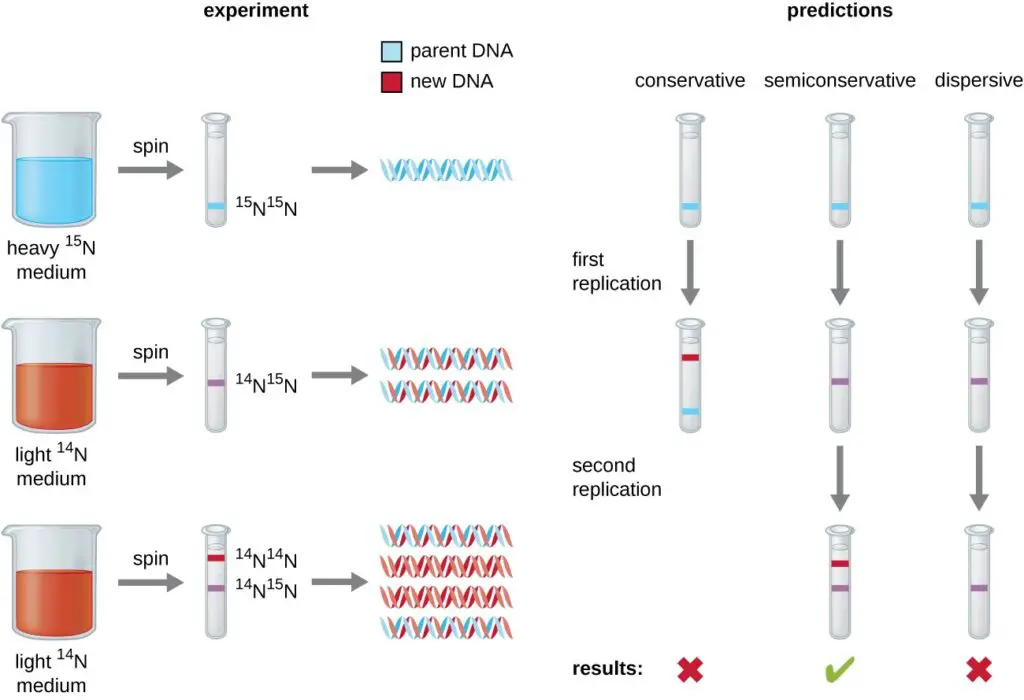
- The process of DNA replication is termed “semiconservative” due to the manner in which DNA molecules are duplicated. This concept was elucidated following the groundbreaking discovery of the double helix structure of DNA by James Watson and Francis Crick in 1953. Their revelation suggested a potential mechanism for DNA replication, where the two strands of the double helix separate and each serves as a template for the synthesis of a new complementary strand.
- In semiconservative replication, when the double helix is unwound, each of the two parental strands acts as a template. As new nucleotides are added, they form complementary base pairs with the template strand. Consequently, each newly synthesized DNA molecule consists of one original (parental) strand and one newly formed strand. This ensures that the replicated DNA retains the original genetic information.
- To validate this model of replication, Matthew Meselson and Franklin Stahl conducted a pivotal experiment in 1958. They cultivated the bacterium Escherichia coli in a medium containing a heavy isotope of nitrogen (15N), which became incorporated into the DNA.
- Subsequently, they shifted the bacteria to a medium with a lighter isotope (14N) and observed the DNA after one and two generations using ultracentrifugation. The results displayed distinct bands representing DNA densities.
- After one generation in the 14N medium, the DNA showed an intermediate density, suggesting that it contained a mix of 15N and 14N. After two generations, two bands were evident: one of intermediate density and one corresponding to 14N DNA.
- These observations confirmed that each new DNA molecule consisted of one parental strand and one newly synthesized strand, thereby validating the semiconservative model of DNA replication.
- In essence, the semiconservative nature of DNA replication ensures the preservation and accurate transmission of genetic information from one generation of cells to the next, with each daughter DNA molecule inheriting one strand from the original DNA and one newly synthesized strand.
Why DNA Synthesis Proceeds in a 5’-3’,Direction and Is Semi Discontinuous?
DNA synthesis is a meticulously orchestrated process that ensures the accurate replication of genetic information. The directionality and manner in which this synthesis occurs are fundamental to its precision.
Directionality of DNA Synthesis (5′-3′ Direction): DNA synthesis always proceeds in the 5′-3′ direction. This is because the enzymes responsible for adding nucleotides, known as DNA polymerases, can only add nucleotides to the free 3′ hydroxyl (OH) group of the growing DNA chain. As the synthesis progresses, nucleotides are sequentially added to this 3′ end, ensuring the elongation of the new strand in the 5′-3′ direction.
Semidiscontinuous Nature of DNA Synthesis: The two strands of the DNA double helix run in antiparallel directions, meaning one strand runs from 5′ to 3′ while the other runs from 3′ to 5′. Given that DNA synthesis can only proceed in the 5′-3′ direction, this poses a challenge when replicating both strands simultaneously as the replication fork progresses.
To address this challenge:
- Leading Strand Synthesis: One strand, termed the “leading strand,” is synthesized continuously in the 5′-3′ direction, moving with the replication fork.
- Lagging Strand Synthesis: The other strand, known as the “lagging strand,” is synthesized discontinuously. This synthesis involves the creation of short DNA segments called Okazaki fragments, named after Reiji Okazaki who discovered them in the 1960s. For each fragment, synthesis starts with a short RNA primer, followed by DNA synthesis in the 5′-3′ direction. However, since this direction is opposite to the movement of the replication fork, the synthesis of the lagging strand occurs in a piecewise manner, producing multiple Okazaki fragments.
Once all the Okazaki fragments are synthesized, they are joined together by the enzyme DNA ligase to form a continuous DNA strand.
In conclusion, the inherent properties of DNA polymerases and the antiparallel nature of DNA strands dictate the 5′-3′ directionality and semidiscontinuous nature of DNA synthesis. This ensures that both strands of the DNA are accurately and efficiently replicated as the replication fork advances.
DNA Replication Mind Map
Which drug inhibits DNA synthesis?
There are several drugs that inhibit DNA synthesis. These drugs can be classified into different categories based on their mechanism of action.
One class of drugs that inhibits DNA synthesis is antimetabolites. These drugs mimic the structure of essential metabolic molecules, such as nucleic acids and proteins, and interfere with their function. Examples of antimetabolites that inhibit DNA synthesis include:
- Methotrexate: This drug inhibits the enzyme dihydrofolate reductase, which is involved in the synthesis of thymidine, one of the building blocks of DNA.
- Azathioprine: This drug inhibits the enzyme inosine monophosphate dehydrogenase, which is involved in the synthesis of guanosine, another building block of DNA.
- Cytarabine: This drug inhibits the enzyme ribonucleotide reductase, which is involved in the synthesis of deoxyribonucleotides, the precursors to DNA.
Other drugs that inhibit DNA synthesis include:
- Cisplatin: This drug forms covalent bonds with DNA, causing DNA strands to cross-link and preventing DNA synthesis.
- Bleomycin: This drug generates reactive oxygen species that damage DNA, leading to DNA strand breaks and inhibiting DNA synthesis.
- Doxorubicin: This drug intercalates into DNA and causes DNA damage, leading to DNA strand breaks and inhibiting DNA synthesis.
It is important to note that these drugs can have serious side effects, and their use is typically limited to specific medical conditions. They should only be used under the supervision of a healthcare professional.
When does dna replication occur?
DNA replication happens in DNA replication occurs during the S (synthesis) period of cell development. In eukaryotic cells that comprise human cells the cell cycle is comprised of distinct phases that include G1 (gap 1) (gap 1), S (synthesis) G2 (gap 2) as well as M (mitosis).
In the S phase in the S phase, cells’ DNA is replicated to prepare for cell division. It is a nitty-gritty procedure that involves unwinding in the structure known as the double helix and the division of DNA strands and the synthesis of complement strands by using already existing DNA strands for models. The result is the creation from two copies DNA molecules, also known as sister chromatids. These are linked by a region known as the centromere.
Following DNA replication in the S phase Cells progress into its G2 phase, in which it prepares for division of the cell. After G2 cells enter the M phase. In this stage, DNA replication is divided equally between two cells via the process known as mitosis.
It’s crucial to understand the fact that replication of DNA is an essential process that happens in all cells that divide to ensure the correct transfer of genetic information from an generation onto the following.
Where does dna replication occur?
DNA replication takes place within the nucleus cells that are eukaryotic. Eukaryotic cells, including the ones found within humans as well as other organisms with multicellularity, possess an individual compartment known as the nucleus in which DNA is kept.
In the nucleus of the cell DNA replication happens in a highly controlled and coordinated way. The process involves unwinding of the double helix, as well as the formation of new strands that complement each other. Specific proteins and enzymes are involved in the process of replication in order to guarantee the precision and efficacy in DNA replication.
In prokaryotic cell types, which comprise bacteria, the process of DNA replication takes place in the cytoplasm because there is no nucleus. Within these cell types, DNA is found as an elongated molecule. replication may occur at one specific location known as the site of replication.
In the end, whether within the nucleus in eukaryotic cells, or the prokaryotic cell cytoplasm DNA replication is an essential procedure that enables the reliable transmission of genetic information another generation.
What is the purpose of dna replication?
The goal for DNA replication is guarantee the accuracy of transfer in genetic info from one generation to the following. DNA replication is a crucial procedure that occurs prior to cell division and serves a variety of functions:
- Genetic Replication: The process of DNA replication makes sure that every daughter cell receives an exact replica that contains the same genetic content. It ensures the accurate transfer of information about genetics from mother cells back to the daughter cell during the process of cell division.
- Growth and development: DNA replication permits cells to divide and grow which allows for the development and maintenance of organs and tissues. In the process of producing two identical duplicates of DNA molecules, DNA replication supplies the genetic material that allows new cells to grow during tissue repair or growth processes.
- Repair and maintenance: DNA replication plays a part in the DNA repair mechanism. If DNA is damaged by external causes or mistakes in replication, cells are able to make use of the unaffected DNA strand to repair the damaged area during the process of replication.
- Genetic Evolution as well as Genetic Diversity: DNA replication is crucial for the creation of genetic diversity and also the development process. Replication that is accurate assures that genetic changes like mutations, or Recombination events, are effectively transmitted to generations afterward.
The main purpose in DNA replication is ensure DNA’s integrity, and to ensure the correct transmission of information genetic which allows for the growth, development and longevity of living organisms. It is a crucial process that is the basis for the transmission of traits as well as the longevity of life.
Quiz Practice
DNA Replication Worksheet
FAQ
What is dna replication?
DNA replication refers to the copying of genetic information from one strand into another strand, which occurs during cell division. This process allows cells to pass copies of genes onto daughter cells. Replication takes place in each cell cycle. During S phase, DNA replicates itself.
dna replication takes place in which phase?
DNA replication occurs during mitosis, when cells divide. During prophase, chromosomes condense and become visible under the microscope. Condensed chromosomes are separated into two daughter cells. The cell then enters prometaphase, telophase, and metaphase. At the end of metaphase, each chromosome has been assigned to one of the new daughter cells.
During dna replication when unwinding of the interwined double helix occurs the enzymes that can break and reseal one strand are called
The enzyme which is used to unwind DNA is the DNA helicase enzyme, which belongs to the class Helicase and is important for all organisms.
It moves along the double-stranded DNA and separates the strands by breaking hydrogen bonds at the expense of energy from ATP.
DNA ligase is a specific type of enzyme, a ligase facilitates the joining of DNA strands together by catalyzing the formation of phosphodiester bonds.
What is the function of rna primer during dna replication?
A primer is a short nucleic acid sequence that provides a starting point for DNA synthesis. In living organisms, primers are short strands of RNA. A primer must be synthesized by an enzyme called primase, which is a type of RNA polymerase, before DNA replication can occur.
why dna replication is called semiconservative?
Replication is called semiconservative because at the time of replication, in each of the two copies of the DNA, one of the strands of DNA is old and conserved and one is newly formed.
When does dna replication takes place?
DNA replication takes place during the Synthesis (S) phase of cell cycle. During S phase, an enzyme called helicase unwinds the DNA strand. The two single DNA strands can then be used as templates to form two identical double DNA strands.
Which enzyme in dna replication has proofreading ability?
DNA polymerase has the proofreading ability.
When does dna replication occur?
DNA replication occurs during the synthesis or S phase of the cell cycle. The cell cycle is the process by which a cell grows and divides into two daughter cells. DNA replication is an essential process that occurs before cell division, in order to ensure that the genetic information of the cell is accurately passed on to the daughter cells.
During DNA replication, the double-stranded DNA molecule is unwound and each strand serves as a template for the synthesis of a new complementary strand. This process is carried out by a group of proteins known as the replication machinery, which includes enzymes called helicases, primases, and polymerases.
The process of DNA replication is highly accurate, but errors can sometimes occur. These errors, known as mutations, can have a variety of effects on the function of the cell and the organism as a whole.
What is dna replication?
DNA replication is the process by which cells produce copies of their DNA before they divide. DNA replication is an essential process that occurs in all living cells and is required for the maintenance of genetic continuity from one generation to the next.
During DNA replication, the double-stranded DNA molecule is unwound and each strand serves as a template for the synthesis of a new complementary strand. This process is carried out by a group of proteins known as the replication machinery, which includes enzymes called helicases, primases, and polymerases.
The process of DNA replication is highly accurate, but errors can sometimes occur. These errors, known as mutations, can have a variety of effects on the function of the cell and the organism as a whole.
Where does dna replication occur?
DNA replication occurs within the cells of living organisms. It is the process by which cells copy their genetic material before cell division.
During DNA replication, the two strands of the double helix are separated, and each strand serves as a template for the synthesis of a new complementary strand. This process occurs in the cell’s nucleus for most eukaryotes (organisms with a nucleus), and in the cytoplasm for some prokaryotes (single-celled organisms without a nucleus).
The process of DNA replication is essential for the continuation of life, as it allows cells to produce copies of themselves and pass on their genetic information to their offspring. It is also important in the study of genetics and molecular biology, as it allows researchers to understand how DNA is organized and how it is passed from one generation to the next.
Why is dna replication important?
DNA replication is an essential process that occurs in all living cells. It is important for several reasons:
It allows cells to produce copies of themselves before cell division: Before a cell divides, it must replicate its DNA so that each daughter cell receives a complete set of genetic instructions. Without DNA replication, cells would not be able to divide and produce offspring.
It ensures the accuracy of genetic information: DNA replication is a highly accurate process, but errors can occasionally occur. These errors, called mutations, can have a variety of effects on an organism, ranging from benign to harmful. However, the accuracy of DNA replication is important because it ensures that the genetic information is passed on accurately from one generation to the next.
It is essential for the continuation of life: DNA replication is a fundamental process that occurs in all living organisms. Without it, life as we know it would not be possible.
It is important in the study of genetics: The process of DNA replication is essential for understanding how DNA is organized and how it is passed from one generation to the next. This knowledge is important in fields such as molecular biology, genetics, and medicine.
What is the purpose of dna replication?
The primary purpose of DNA replication is to produce copies of the genetic material contained in DNA before cell division. This ensures that each daughter cell receives a complete set of genetic instructions and is able to function properly.
DNA replication is essential for the continuation of life, as it allows cells to produce offspring and pass on their genetic information to future generations. It also ensures the accuracy of genetic information, as errors that occur during DNA replication can have a variety of effects on an organism.
In addition to its role in the continuation of life, DNA replication is also important in the study of genetics and molecular biology, as it allows researchers to understand how DNA is organized and how it is passed from one generation to the next.
