Table of Contents
What are Gram-negative bacteria?
Gram-negative bacteria are a distinct group of bacteria that exhibit specific characteristics in terms of their cell structure and staining properties. These bacteria do not retain the crystal violet stain used in the Gram staining method, which is a common technique for differentiating bacteria. The key features of Gram-negative bacteria include their unique cell envelope structure and the presence of an outer membrane.
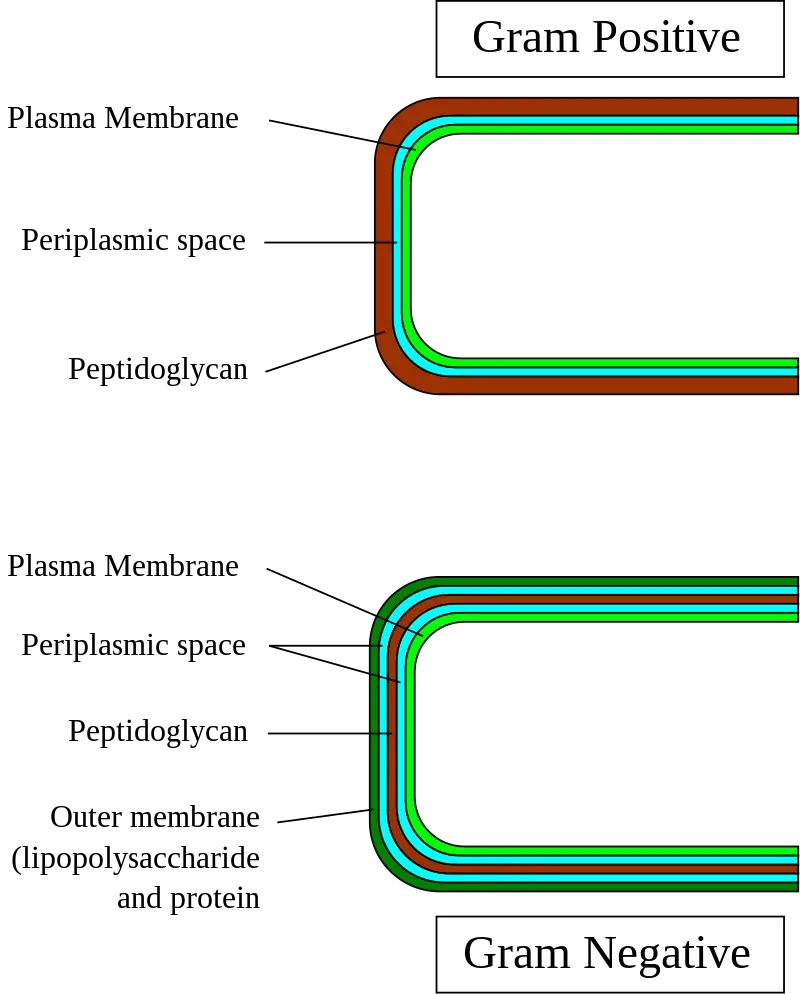
The cell envelope of Gram-negative bacteria consists of a thin layer of peptidoglycan, a type of polymer composed of sugars and amino acids, which is located between an inner cytoplasmic cell membrane and an outer membrane. This outer membrane is a defining characteristic of Gram-negative bacteria and is absent in Gram-positive bacteria. The outer membrane of Gram-negative bacteria contains lipopolysaccharides (LPS) in its outer leaflet, which can trigger a toxic reaction when the bacteria are lysed by immune cells.
Gram-negative bacteria are found in diverse environments that support life. They can be encountered in soil, water, plants, animals, and even the human body. Some well-known examples of Gram-negative bacteria include Escherichia coli (E. coli), Pseudomonas aeruginosa, Chlamydia trachomatis, and Yersinia pestis (the causative agent of the bubonic plague).
One of the challenges posed by Gram-negative bacteria is their resistance to certain antibiotics and immune defenses. The outer membrane acts as a barrier, protecting the bacteria from the entry of various substances, including antibiotics and detergents. Furthermore, the lipopolysaccharide component of the outer membrane can cause a toxic reaction when the bacteria are destroyed, leading to severe symptoms such as low blood pressure, respiratory failure, and septic shock.
To combat infections caused by Gram-negative bacteria, specific classes of antibiotics have been developed. These antibiotics are designed to target the unique characteristics of Gram-negative bacteria and include aminopenicillins, ureidopenicillins, cephalosporins, quinolones, and carbapenems. Some antibiotics, such as aminoglycosides, monobactams (e.g., aztreonam), and ciprofloxacin, specifically target Gram-negative organisms.
In summary, Gram-negative bacteria are a diverse group of bacteria characterized by their cell envelope structure, which includes a thin layer of peptidoglycan and an outer membrane. They can be found in various environments and include both harmless and pathogenic species. Understanding the unique features of Gram-negative bacteria is important for developing effective treatments and combating infections caused by these organisms.
Definition of Gram-negative bacteria
Gram-negative bacteria are a group of bacteria that do not retain the crystal violet stain in the Gram staining method, appearing pink or red instead. They have a unique cell envelope structure consisting of a thin peptidoglycan layer surrounded by an outer membrane. Gram-negative bacteria can be found in diverse environments and include both harmless and pathogenic species. They are characterized by their resistance to certain antibiotics and their ability to cause severe infections.
Characteristics of Gram-negative bacteria
Gram-negative bacteria possess several characteristic features, including:
- Inner cell membrane: Gram-negative bacteria have an inner cell membrane, also known as the cytoplasmic membrane, which separates the cytoplasm from the external environment.
- Thin peptidoglycan layer: Compared to gram-positive bacteria, gram-negative bacteria have a thin peptidoglycan layer in their cell wall.
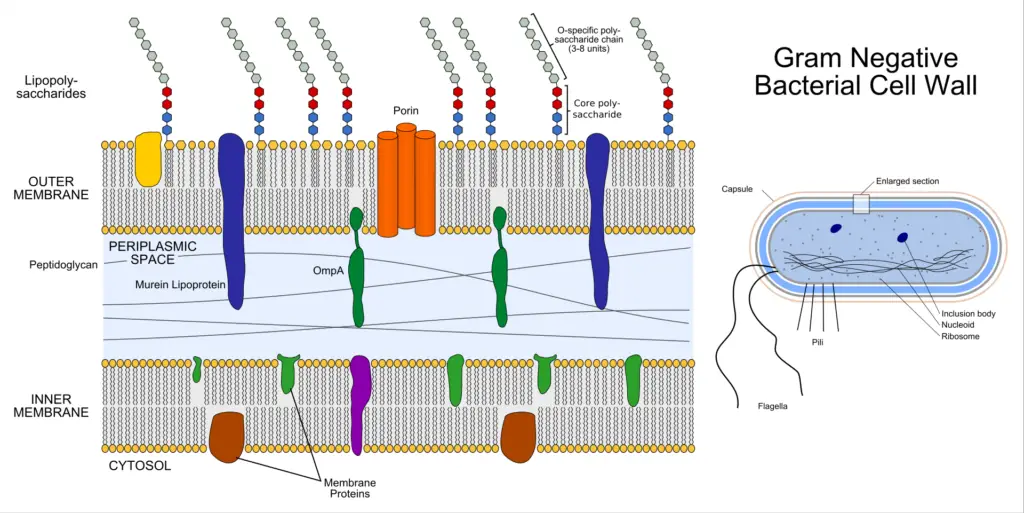
- Outer membrane: Gram-negative bacteria have an outer membrane that contains lipopolysaccharides (LPS) in its outer leaflet and phospholipids in the inner leaflet. The outer membrane provides an additional protective barrier.
- Porins: Outer membrane of gram-negative bacteria contains porins, which are protein channels that act as pores, allowing the passage of specific molecules into the periplasmic space.
- Periplasmic space: Between the outer membrane and the inner cell membrane, gram-negative bacteria have a space called the periplasmic space. This space is filled with a concentrated gel-like substance known as periplasm, which contains various enzymes and proteins.
- S-layer: The S-layer, a protein layer, is directly attached to the outer membrane in gram-negative bacteria.
- Flagella: If present, the flagella of gram-negative bacteria have four supporting rings, as opposed to the two rings found in gram-positive bacteria.
- Teichoic acids: Gram-negative bacteria do not possess teichoic acids or lipoteichoic acids, which are present in gram-positive bacteria.
- Lipoproteins: Lipoproteins are attached to the polysaccharide backbone of gram-negative bacteria.
- Braun’s lipoprotein: Some gram-negative bacteria contain Braun’s lipoprotein, which acts as a covalent bond between the outer membrane and the peptidoglycan layer.
- Spore formation: Most gram-negative bacteria do not form spores, although there are a few exceptions.
The unique cell envelope structure and composition of gram-negative bacteria contribute to their distinct characteristics and behaviors.
Gram-negative bacteria shapes
Gram-negative bacteria exhibit a diverse range of shapes when observed under a microscope. While the most commonly known shapes include rods (bacillus), cocci, and spirals, there are also some special shapes observed among Gram-negative bacteria. Here are a few examples:
- Rod/bacillus shaped: Escherichia coli is a well-known Gram-negative bacterium that exhibits a rod-shaped morphology.
- Coccobacillus: This shape is a combination of both cocci (spherical) and bacilli (rod-shaped) forms. Hemophilus influenza is an example of a Gram-negative bacterium that displays a coccobacillus shape.
- Streptobacillus: Streptobacilli are rod-shaped bacteria that are connected together in chains. An example of a Gram-negative bacterium with this shape is Streptobacillus moniliformis.
- Trichome shape: Some Gram-negative bacteria form trichomes, which are series of rod-shaped cells arranged in a columnar form and may be enclosed in a sheath.
- Spiral shaped bacteria (Spirochaetes): Spirochaetes are Gram-negative bacteria that exhibit a spiral shape. Examples of spiral-shaped Gram-negative bacteria include Chlamydia trachomatis and Treponema pallidum.
- Filamentous shaped: Certain Gram-negative bacteria have a filament-like shape. For instance, species of Nocardia display filamentous morphology.
These variations in shape among Gram-negative bacteria contribute to their diverse characteristics and adaptations to different environments.
Cell wall of Gram-negative bacteria
The cell wall of Gram-negative bacteria is characterized by its complexity compared to that of Gram-positive bacteria. It consists of multiple layers, including a thin layer of peptidoglycan and a thick outer membrane. Here are some key points about the cell wall of Gram-negative bacteria:
- Peptidoglycan layer: Gram-negative bacteria have a relatively thin peptidoglycan layer, ranging from 2 to 7 nanometers in thickness. Peptidoglycan is a mesh-like structure composed of sugars and amino acids that provides structural support to the bacterial cell wall.
- Outer membrane: The outer membrane is a unique feature of Gram-negative bacteria and is absent in Gram-positive bacteria. It is a lipid bilayer consisting of phospholipids in the inner leaflet and lipopolysaccharides (LPS) in the outer leaflet. The outer membrane acts as a barrier and provides protection against certain chemicals and antimicrobial agents.
- Periplasmic space: Between the inner cell membrane and the outer membrane lies the periplasmic space, which is larger in Gram-negative bacteria compared to Gram-positive bacteria. The periplasmic space contains a gel-like substance called periplasm, which contains various enzymes, transport proteins, and other molecules important for cellular functions.
- Lipopolysaccharides (LPS): The outer leaflet of the outer membrane in Gram-negative bacteria contains lipopolysaccharides (LPS). LPS consists of three components: lipid A, core polysaccharide, and O antigen. LPS plays a crucial role in maintaining the structural integrity of the outer membrane and can also induce an immune response in the host organism.
- Porins: The outer membrane of Gram-negative bacteria contains porins, which are protein channels that allow the passage of certain molecules across the membrane. These porins act as gatekeepers, regulating the entry and exit of molecules into the periplasmic space.
The complex structure of the cell wall in Gram-negative bacteria provides them with unique properties and functions. It plays a critical role in maintaining cell shape, protecting the cell from external stresses, and facilitating interactions with the environment.
The Periplasmic space
The periplasmic space is a unique compartment found in Gram-negative bacteria, located between the inner cell membrane and the outer membrane. It plays a crucial role in various cellular processes and is involved in acquiring nutrients, maintaining cell integrity, and modifying potentially harmful substances. Here are some key aspects of the periplasmic space:
- Nutrient acquisition: The periplasmic space contains proteins known as binding proteins that actively participate in the uptake of nutrients. These binding proteins bind to specific molecules outside the bacterial cell, such as sugars, amino acids, and ions, and transport them into the periplasmic space. From there, the nutrients can be further processed and transported into the cytoplasm for utilization by the bacterium.
- Hydrolytic enzymes: Within the periplasmic space, Gram-negative bacteria house hydrolytic enzymes that can degrade various macromolecules, including nucleic acids and phosphorylated molecules. These enzymes aid in the breakdown of complex substrates, allowing the bacterium to extract essential components for metabolism.
- Peptidoglycan synthesis: The periplasmic space is also involved in the synthesis and modification of peptidoglycan, a critical component of the bacterial cell wall. Enzymes within the periplasmic space contribute to the assembly of peptidoglycan by catalyzing the formation of peptide cross-links and modifying existing peptidoglycan structures.
- Detoxification: Toxic substances encountered by the bacterium, such as heavy metals or antibiotics, can be modified or neutralized within the periplasmic space. Enzymes in this compartment can chemically modify toxic compounds to reduce their harmful effects on the bacterial cell.
Overall, the periplasmic space serves as a dynamic and versatile region in Gram-negative bacteria. It houses proteins involved in nutrient acquisition, hydrolytic activities, peptidoglycan synthesis, and detoxification processes. The presence of this space allows for compartmentalization of specific functions, enhancing the efficiency and adaptability of the bacterial cell.
Peptidoglycan
Peptidoglycan is a crucial component of the cell wall in bacteria, including Gram-negative bacteria. It forms a mesh-like structure that provides strength and rigidity to the bacterial cell wall. Here are some key points about peptidoglycan:
- Composition: Peptidoglycan is primarily composed of long chains of alternating N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM) sugars. These sugar chains are cross-linked by short peptides, forming a strong and interconnected network.
- Structural role: Peptidoglycan serves as a structural scaffold, giving the bacterial cell wall its shape and integrity. It provides strength and protection against osmotic pressure changes and mechanical stress.
- Thickness in Gram-negative bacteria: In Gram-negative bacteria, the peptidoglycan layer is relatively thin compared to Gram-positive bacteria. It is located between the inner cell membrane and the outer membrane, making up about 5-10% of the bacterial cell’s dry weight. Some Gram-negative bacteria, like Escherichia coli, have a peptidoglycan layer that is approximately 2 nanometers thick, consisting of 2-3 sheets of peptidoglycan.
- Cross-linking: The peptidoglycan chains are cross-linked through peptide bridges, which are formed by the transpeptidation reaction catalyzed by enzymes called penicillin-binding proteins (PBPs). The cross-linking of peptidoglycan provides additional strength and stability to the cell wall.
- Target of antibiotics: Peptidoglycan synthesis and cross-linking are essential processes for bacterial cell wall formation. Consequently, many antibiotics, such as penicillins and cephalosporins, target the enzymes involved in peptidoglycan synthesis or disrupt the cross-linking process, leading to weakened cell walls and bacterial cell death.
- Role in bacterial growth and division: Peptidoglycan is essential for bacterial growth and division. During cell growth, new peptidoglycan material is synthesized and inserted into the existing cell wall, allowing the bacterium to expand and elongate. During cell division, peptidoglycan synthesis and remodeling play a critical role in separating the daughter cells.
In summary, peptidoglycan is a vital component of the cell wall in Gram-negative bacteria. It provides structural support, contributes to the overall integrity of the cell wall, and plays a crucial role in bacterial growth and division. Understanding the composition and function of peptidoglycan is essential for developing strategies to target bacterial cell walls for therapeutic purposes.
The Outer Membrane and the Lipopolysaccharides
The outer membrane and lipopolysaccharides (LPS) are important components of Gram-negative bacteria. Here are some key points about the outer membrane and LPS:
- Outer membrane structure: The outer membrane is located outside the thin peptidoglycan layer in Gram-negative bacteria. It consists of various components, including Braun’s lipoprotein, which covalently binds the outer membrane to the peptidoglycan layer. Adhesion sites on the outer membrane play a role in cell contact and membrane fusion.
- Lipopolysaccharides (LPS): The outer membrane is predominantly composed of LPS, which are large complex molecules consisting of lipids and carbohydrates. LPS is composed of three units: Lipid A, core polysaccharides, and the O side chain. Lipid A contains fatty acids, glucosamine sugars, and pyrophosphate, and it contributes to the toxic properties of LPS as endotoxins. The O side chain, also known as the O antigen, extends outwardly from the core and is composed of sugars that vary between bacterial strains. These O antigens play a role in evading antibody responses.
- Protection and stability: LPS in the outer membrane serves to protect the cell wall from external attacks, such as antibiotics, bile salts, and other toxic substances. Additionally, LPS imparts a negative charge to the cell surface, which helps stabilize the membrane structure.
- Antibiotic permeability: The outer membrane is selectively permeable due to the presence of porin proteins. These porins form channels that allow the entry of small molecules, such as glucose, into the bacterial cell. However, larger molecules like Vitamin B12 require specific carrier proteins for transport across the outer membrane.
- Prevention of component loss: The outer membrane plays a role in preventing the loss of components, particularly from the periplasmic space. It acts as a barrier to retain substances within the bacterial cell.
In summary, the outer membrane and lipopolysaccharides are crucial components of Gram-negative bacteria. The outer membrane provides structural support, protection against external threats, and selective permeability. Lipopolysaccharides, specifically Lipid A and the O antigen, contribute to the toxic properties of Gram-negative bacteria and play a role in immune evasion. Understanding the structure and function of the outer membrane and LPS is important for studying bacterial pathogenesis, antibiotic resistance, and developing strategies to combat Gram-negative bacterial infections.
Examples of Gram-negative bacteria and their pathologies and clinical significance
Gram-negative bacteria encompass a wide range of species, some of which are known for causing significant human infections and diseases. Here are examples of Gram-negative bacteria, along with their pathologies and clinical significance:
- Neisseria gonorrhoeae:
- Genitourinary tract infections: Infections in males can lead to urethritis with purulent discharge and painful urination. In females, infections can affect the vagina and endocervix, leading to symptoms such as purulent discharge, painful urination, salpingitis, pelvic inflammatory disease (PID), and fibrosis.
- Renal infection: In males, the bacteria can cause renal infection, resulting in constipation, painful defecation, and purulent discharge.
- Other infections: Neisseria gonorrhoeae can also cause pharyngitis, ophthalmia neonatorum in newborns, and disseminated infections characterized by fever, purulent arthritis, and skin pustules.
- Neisseria meningitidis:
- Meningitis: The bacteria can cause meningitis, which is characterized by high fever, severe headaches, joint aches, and a petechial and/or purpuric rash.
- Meningococcemia: In severe cases, Neisseria meningitidis can invade the bloodstream, leading to meningococcemia, septicemia, and shock, including the Waterhouse-Friderichsen syndrome.
- Escherichia coli:
- Intestinal diseases: Different strains of E. coli, such as enterotoxigenic, enteropathogenic, enterohemorrhagic, enteroinvasive, and enteroaggregative, can cause various forms of intestinal diseases associated with diarrhea (watery or/and bloody).
- Extraintestinal diseases: E. coli can cause urinary tract infections (cystitis and pyelonephritis), neonatal meningitis, and nosocomial-acquired infections, including sepsis/bacteremia, endotoxic shock, and pneumonia.
- Salmonella spp:
- Enteric and Typhoid fever: Infections with Salmonella can lead to fevers, abdominal pain, chills, sweats, headache, anorexia, weakness, sore throat, and either diarrhea or constipation.
- Gastroenteritis (salmonellosis): This form of infection presents with symptoms such as nausea, vomiting, and non-bloody diarrhea.
- Bacteremia and other complications: Salmonella infections can lead to bacteremia, abdominal infections, osteomyelitis, and septic arthritis.
- Campylobacter jejuni:
- Intestinal and extraintestinal diseases: Infections with Campylobacter jejuni can cause systemic symptoms such as fever, headache, myalgia, abdominal cramping, and diarrhea (which may or may not be bloody). It is a common cause of traveler’s diarrhea and can mimic appendicitis without inflammation of the appendix.
- Vibrio cholerae:
- Cholera: Infection with Vibrio cholerae leads to cholera, characterized by profuse watery diarrhea and massive loss of fluid and electrolytes from the body.
- Helicobacter pylori:
- Gastric diseases: Helicobacter pylori is associated with acute gastritis, superficial gastritis with epigastric discomfort, duodenal ulcers, gastric ulcers, and, in persistent cases, mucosa-associated lymphoid tumors.
- Klebsiella pneumoniae:
- Urinary tract infections (UTI) and nosocomial bacteremia: Klebsiella pneumoniae is known for causing UTIs and nosocomial-acquired bacteremia.
- Pseudomonas aeruginosa:
- Opportunistic infections: Pseudomonas aeruginosa causes nosocomial infections in wounded patients, often entering the body through catheters and respirators. It can cause keratitis, endophthalmitis, skin wound infections, respiratory tract infections (including pneumonia), gastrointestinal infections with diarrhea, necrotic enterocolitis in infants, and systemic infections in hospitalized patients.
These examples illustrate the diverse pathologies and clinical significance associated with Gram-negative bacteria. Understanding the specific bacteria involved in infections is crucial for appropriate diagnosis, treatment, and prevention strategies.
Antimicrobial agents For Gram-negative bacteria
Antimicrobial agents play a crucial role in combating infections caused by Gram-negative bacteria. These agents, commonly referred to as antibiotics, work by targeting specific mechanisms within bacterial cells to inhibit their growth and replication. Here are examples of antimicrobial agents commonly used against Gram-negative bacteria:
- Cephalosporin (e.g., ceftriaxone):
- Mode of action: Disrupts the bacterial cell by binding to penicillin-binding proteins and enzymes involved in peptidoglycan synthesis.
- Bacterial agents: Neisseria gonorrhoeae, Neisseria meningitidis, Pseudomonas aeruginosa.
- Tetracycline (e.g., doxycycline):
- Mode of action: Inhibits protein synthesis by preventing the elongation of polypeptides at 30s ribosomes.
- Bacterial agents: Neisseria gonorrhoeae, Streptogramins.
- β-lactam (e.g., Penicillin G):
- Mode of action: Inhibits cell wall synthesis by disrupting penicillin-binding proteins and enzymes involved in peptidoglycan synthesis.
- Bacterial agents: Neisseria meningitidis, Pseudomonas aeruginosa.
- Rifampin (rifamycin):
- Mode of action: Inhibits nucleic acid synthesis by preventing transcription through binding to DNA-dependent RNA polymerase.
- Bacterial agents: Neisseria meningitidis, Escherichia coli.
- Macrolides (e.g., erythromycin, azithromycin, clarithromycin):
- Mode of action: Inhibits bacterial protein synthesis by preventing elongation of polypeptides at 50s ribosomes.
- Bacterial agents: Neisseria gonorrhoeae, Campylobacter jejuni, Shigella dysenteriae, Helicobacter pylori, Pseudomonas aeruginosa.
- Quinolones (e.g., fluoroquinolones, ciprofloxacin):
- Mode of action: Inhibits nucleic acid synthesis by binding to the alpha-subunit of DNA gyrase.
- Bacterial agents: Escherichia coli, Salmonella typhi/paratyphi, Campylobacter jejuni, Shigella dysenteriae, Pseudomonas aeruginosa.
- Aminoglycosides:
- Mode of action: Inhibit protein synthesis by prematurely producing aberrant peptide chains from 30s ribosomes.
- Bacterial agent: Escherichia coli (localized and systemic infections).
- Sulfonamides (e.g., sulfamethoxazole) and Trimethoprim:
- Mode of action: Sulfonamides inhibit dihydropteroate synthase, while trimethoprim inhibits dihydrofolate reductase, both disrupting folic acid synthesis.
- Bacterial agent: Escherichia coli (UTIs and systemic diseases).
These examples demonstrate the diverse mechanisms of action used by antimicrobial agents to target Gram-negative bacteria and highlight the importance of selecting the appropriate antibiotic based on the specific bacterial agent and the nature of the infection.
Non proteobacteria: General characteristics with suitable examples
Non-proteobacteria refer to a group of Gram-negative bacteria that are distinct from the Proteobacteria phylum. Here are some general characteristics of non-proteobacteria along with suitable examples:
- Cyanobacteria: Cyanobacteria are photosynthetic bacteria capable of oxygenic photosynthesis. They play a crucial role in oxygen production and are commonly found in aquatic environments. Examples include Anabaena, Nostoc, and Spirulina.
- Spirochetes: Spirochetes are characterized by their spiral or corkscrew-like shape. They are motile bacteria and often exhibit unique modes of locomotion. The best-known example is Treponema pallidum, the causative agent of syphilis.
- Chlamydiae: Chlamydiae are obligate intracellular bacteria that can cause a variety of infections in humans and animals. They have a unique developmental cycle and are associated with diseases such as chlamydia, trachoma, and pneumonia. Chlamydia trachomatis is one of the most notable species.
- Bacteroidetes: Bacteroidetes are a diverse group of bacteria found in various environments, including the human gut. They play an essential role in digestion and are involved in the degradation of complex carbohydrates. Examples include Bacteroides fragilis and Prevotella.
- Fusobacteria: Fusobacteria are anaerobic bacteria commonly found in the oral cavity and gastrointestinal tract. They are associated with various infections, such as periodontal diseases and Lemierre’s syndrome. Fusobacterium nucleatum is a well-known species.
- Planctomycetes: Planctomycetes are unique bacteria with complex cell structures. They exhibit features typically found in eukaryotes, such as compartmentalization and endocytosis-like processes. Some examples include Planctomyces and Gemmata.
- Spiroplasma: Spiroplasma are helical bacteria that lack a cell wall. They are often associated with insects and can be both pathogenic and symbiotic. Spiroplasma citri is known to cause citrus stubborn disease in plants.
- Deinococcus-Thermus: Deinococcus-Thermus bacteria are highly resistant to extreme conditions, including radiation and desiccation. They are commonly found in environments such as hot springs. Thermus aquaticus is notable for its heat-resistant DNA polymerase, Taq polymerase, widely used in polymerase chain reaction (PCR) technology.
These examples represent some of the diverse non-proteobacteria groups. Each group exhibits unique characteristics, ecological roles, and potential interactions with their environment and hosts.
Alpha proteobacteria: General characteristics with suitable examples
Alpha-proteobacteria are a group of Gram-negative bacteria belonging to the Proteobacteria phylum. They are diverse and have a wide range of characteristics. Here are some general characteristics of alpha-proteobacteria along with suitable examples:
- Intracellular symbionts: Many alpha-proteobacteria have developed intimate associations with eukaryotic hosts and live as intracellular symbionts. An example is Wolbachia, which infects a wide range of arthropods and is known for its ability to manipulate host reproduction.
- Nitrogen-fixing bacteria: Alpha-proteobacteria include several nitrogen-fixing bacteria that convert atmospheric nitrogen into a usable form. Rhizobium species, for instance, form symbiotic associations with leguminous plants, where they establish nodules on plant roots and provide them with nitrogen.
- Plant pathogens: Some alpha-proteobacteria can cause diseases in plants. Agrobacterium tumefaciens is a well-known example. It is responsible for crown gall disease, where it transfers a segment of its DNA (T-DNA) into the plant genome, leading to the formation of tumor-like growths.
- Free-living bacteria: Alpha-proteobacteria also include free-living bacteria found in various environments. They can be found in soil, water, and marine ecosystems. One example is Caulobacter crescentus, which has a distinct life cycle with two different cell types and plays a role in freshwater ecosystems.
- Rhodospirillaceae: This family of alpha-proteobacteria consists of phototrophic bacteria that can carry out photosynthesis. They use various pigments, including bacteriochlorophyll, and can be found in diverse habitats such as freshwater, marine environments, and even the gastrointestinal tracts of animals.
- Rickettsiales: Rickettsiales are obligate intracellular bacteria that can cause diseases in humans and other animals. They include important pathogens such as Rickettsia species, which are responsible for diseases like Rocky Mountain spotted fever and typhus.
- Caulobacterales: Caulobacterales are bacteria known for their unique cell cycle and development. They have a stalked and a non-stalked cell type, and their life cycle involves asymmetric cell division. Caulobacter spp. are often used as model organisms for studying bacterial cell biology.
These examples represent some of the diverse groups within the alpha-proteobacteria. They exhibit a range of ecological roles, including symbiosis, nitrogen fixation, pathogenesis, and environmental adaptation. Alpha-proteobacteria play significant roles in various ecosystems and have both beneficial and harmful interactions with their hosts.
Beta proteobacteria: General characteristics with suitable examples
Beta-proteobacteria are a group of Gram-negative bacteria belonging to the Proteobacteria phylum. They exhibit diverse characteristics and occupy various ecological niches. Here are some general characteristics of beta-proteobacteria along with suitable examples:
- Nutrient cycling: Many beta-proteobacteria play important roles in nutrient cycling, particularly in the nitrogen cycle. They can oxidize or reduce nitrogen compounds, contributing to the conversion of ammonia to nitrite or nitrate, and vice versa. An example is Nitrosomonas, which is involved in the oxidation of ammonia to nitrite in the process of nitrification.
- Aquatic bacteria: Beta-proteobacteria can be commonly found in aquatic environments such as freshwater and marine ecosystems. They are often associated with the degradation of organic matter. For instance, members of the genus Burkholderia are frequently found in water and soil environments, and some species are involved in the breakdown of complex organic compounds.
- Symbiotic relationships: Some beta-proteobacteria form symbiotic associations with plants or animals. An example is the genus Bordetella, which includes species that are pathogens causing respiratory diseases in humans and animals. Bordetella pertussis, the causative agent of whooping cough, colonizes the respiratory tract and establishes a symbiotic relationship with the host.
- Pathogens: Beta-proteobacteria include several pathogenic species that can cause diseases in humans, animals, and plants. The genus Neisseria comprises pathogenic species such as Neisseria meningitidis, the causative agent of meningitis and meningococcal septicemia, and Neisseria gonorrhoeae, which causes the sexually transmitted infection gonorrhea.
- Environmental adaptation: Beta-proteobacteria exhibit adaptability to diverse environmental conditions. They can thrive in environments with low nutrient availability, such as groundwater and soil. Some species of the genus Cupriavidus, formerly known as Ralstonia, have the ability to degrade a wide range of organic pollutants, making them important in bioremediation processes.
- Iron-oxidizing bacteria: Beta-proteobacteria include iron-oxidizing bacteria that are involved in the oxidation of ferrous iron (Fe2+) to ferric iron (Fe3+). These bacteria play a crucial role in iron cycling in aquatic and terrestrial environments. Acidithiobacillus ferrooxidans is an example of an iron-oxidizing beta-proteobacterium.
- Biotechnological applications: Beta-proteobacteria have been utilized in various biotechnological applications. For instance, members of the genus Burkholderia have been used for the production of antibiotics, enzymes, and bioplastics.
These examples demonstrate the diversity and ecological significance of beta-proteobacteria. They exhibit various metabolic capabilities, including nutrient cycling, pathogenesis, and environmental adaptation. Beta-proteobacteria play important roles in ecosystem processes and have both beneficial and detrimental impacts on humans, animals, and the environment.
Gamma proteobacteria: General characteristics with suitable examples
Gamma-proteobacteria are a diverse group of Gram-negative bacteria belonging to the Proteobacteria phylum. They exhibit a wide range of characteristics and include many important and well-known bacterial species. Here are some general characteristics of gamma-proteobacteria along with suitable examples:
- Ecological diversity: Gamma-proteobacteria have a broad ecological distribution and can be found in various environments such as soil, water, and the gastrointestinal tracts of animals. They display adaptability to different ecological niches and play important roles in nutrient cycling, decomposition of organic matter, and symbiotic associations.
- Pathogens: Many important human and animal pathogens are classified as gamma-proteobacteria. These bacteria possess various virulence factors and mechanisms that enable them to cause diseases. Examples include:
- Escherichia coli: Some strains of E. coli, such as enterohemorrhagic E. coli (EHEC) and enterotoxigenic E. coli (ETEC), can cause gastrointestinal infections, while other strains are associated with urinary tract infections and other extraintestinal infections.
- Salmonella: Salmonella species are responsible for salmonellosis, a foodborne illness characterized by symptoms like diarrhea, abdominal cramps, and fever.
- Vibrio cholerae: This bacterium causes cholera, a severe diarrheal disease with the potential for outbreaks and epidemics in areas with poor sanitation.
- Pseudomonas aeruginosa: P. aeruginosa is an opportunistic pathogen that can cause infections in immunocompromised individuals, particularly those with cystic fibrosis, burn wounds, or compromised respiratory systems.
- Symbiotic associations: Gamma-proteobacteria form symbiotic relationships with various organisms. One notable example is the genus Symbiobacterium, which lives in the intestines of certain insects and plays a role in the digestion of cellulose.
- Nitrogen fixation: Some gamma-proteobacteria have the ability to fix atmospheric nitrogen into a usable form, contributing to nitrogen cycling and availability in the environment. One well-known example is the genus Azotobacter, which can fix nitrogen in free-living conditions.
- Bioremediation: Gamma-proteobacteria are involved in bioremediation processes, where they degrade or detoxify pollutants in the environment. Pseudomonas species, such as Pseudomonas putida and Pseudomonas fluorescens, are widely used for bioremediation due to their metabolic versatility and ability to degrade various organic compounds.
- Industrial and commercial importance: Some gamma-proteobacteria have industrial and commercial applications. For instance, the species Escherichia coli is extensively used in biotechnology and molecular biology as a host organism for recombinant DNA technology and protein production.
- Diversity of metabolic capabilities: Gamma-proteobacteria exhibit a wide range of metabolic capabilities, including the ability to utilize diverse carbon and energy sources. They can be aerobic, facultative anaerobic, or anaerobic, displaying metabolic versatility that contributes to their ecological success.
These examples highlight the diverse nature and significance of gamma-proteobacteria. They encompass a wide range of ecological roles, including pathogens, symbionts, and environmental contributors. Gamma-proteobacteria have both beneficial and detrimental effects on human health, ecosystem functioning, and industrial applications.
Delta proteobacteria: General characteristics with suitable examples
Delta-proteobacteria are a diverse group of Gram-negative bacteria belonging to the Proteobacteria phylum. They encompass a wide range of organisms with distinct characteristics and ecological roles. Here are some general characteristics of delta-proteobacteria along with suitable examples:
- Predominantly anaerobic: Many delta-proteobacteria are anaerobic or facultatively anaerobic, meaning they can survive and thrive in environments with little to no oxygen. They often inhabit oxygen-depleted habitats such as sediments, mud, and gastrointestinal tracts.
- Sulfate-reducing bacteria: One of the most well-known and ecologically significant groups within the delta-proteobacteria are the sulfate-reducing bacteria (SRB). These bacteria obtain energy by reducing sulfate (SO42-) to hydrogen sulfide (H2S), playing a vital role in sulfur and carbon cycles. Examples of sulfate-reducing bacteria include:
- Desulfovibrio: This genus contains several species of sulfate-reducing bacteria that can be found in diverse environments such as marine sediments, freshwater systems, and animal intestines.
- Desulfobacter: These bacteria are commonly found in anaerobic environments and are involved in the degradation of organic compounds.
- Ecological importance: Delta-proteobacteria exhibit ecological importance through their participation in nutrient cycling and their interactions with other organisms. They play roles in anaerobic decomposition processes, carbon cycling, and the breakdown of organic matter.
- Geobacteraceae family: The Geobacteraceae family is a notable group within the delta-proteobacteria. It includes the genus Geobacter, which has garnered attention for its ability to transfer electrons to various electron acceptors, including metals and electrodes. Geobacter species are involved in processes such as bioremediation of organic and metal contaminants and the generation of electricity in microbial fuel cells.
- Myxobacteria: Another group of delta-proteobacteria includes the myxobacteria. These bacteria are known for their unique social behaviors, forming multicellular structures called fruiting bodies when conditions are unfavorable. Myxobacteria have complex life cycles and exhibit gliding motility. Examples include the genus Myxococcus, which can be found in soil habitats and are known for their ability to produce a wide array of secondary metabolites.
- Bdellovibrio: Bdellovibrio is a genus of delta-proteobacteria that is intriguing due to its predatory nature. These bacteria are parasitic and prey upon other Gram-negative bacteria, entering their periplasmic space and utilizing their host’s resources. Bdellovibrio has potential applications in controlling bacterial pathogens.
- Desulfarculaceae family: The Desulfarculaceae family consists of sulfate-reducing bacteria that are involved in sulfur and carbon cycling. They can be found in various environments, including freshwater sediments, hot springs, and hydrothermal vents.
The examples provided highlight the diverse characteristics and ecological roles of delta-proteobacteria. They encompass sulfate-reducing bacteria, predatory bacteria, and those involved in biogeochemical cycles. Delta-proteobacteria contribute significantly to anaerobic environments, nutrient cycling, and ecosystem functioning.
Epsilon proteobacteria: General characteristics with suitable examples
Epsilon-proteobacteria is a class of Gram-negative bacteria within the Proteobacteria phylum. They are a relatively small group but exhibit unique characteristics and diverse ecological roles. Here are some general characteristics of epsilon-proteobacteria along with suitable examples:
- Microaerophilic or microaerobic: Epsilon-proteobacteria are typically microaerophilic, meaning they thrive in environments with low oxygen levels. They can be found in habitats such as marine sediments, hydrothermal vents, and the gastrointestinal tracts of animals.
- Helical or curved shape: Many epsilon-proteobacteria have a helical or curved cell shape, which aids in their movement and colonization of specific environments.
- Pathogenicity: Several epsilon-proteobacteria are known to be human pathogens and are associated with causing diseases. They possess adaptations that allow them to colonize and survive in host tissues. Examples of pathogenic epsilon-proteobacteria include:
- Helicobacter pylori: H. pylori is a well-known epsilon-proteobacterium that colonizes the human stomach and is associated with gastric ulcers, gastritis, and stomach cancer. It has unique mechanisms to survive in the acidic environment of the stomach.
- Campylobacter jejuni: C. jejuni is a common cause of bacterial gastroenteritis worldwide. It is often transmitted through contaminated food and water, leading to symptoms such as diarrhea, abdominal pain, and fever.
- Sulfur metabolism: Some epsilon-proteobacteria are involved in sulfur metabolism. They can utilize sulfur compounds as energy sources or electron acceptors. For example:
- Sulfurimonas: Sulfurimonas species are found in deep-sea hydrothermal vents and participate in sulfur oxidation processes, playing a role in the sulfur cycle.
- Deep-sea hydrothermal vents: Epsilon-proteobacteria are often found in extreme environments such as deep-sea hydrothermal vents. These environments are characterized by high temperatures, high pressure, and chemically rich conditions. Epsilon-proteobacteria, including genera such as Nautilia and Caminibacter, have been identified in hydrothermal vent communities and are involved in various geochemical processes.
- Chemoautotrophy: Some epsilon-proteobacteria are chemoautotrophs, meaning they obtain energy from the oxidation of inorganic compounds and use carbon dioxide as their carbon source. They play important roles in the cycling of sulfur and other elements in marine and deep-sea environments.
Epsilon-proteobacteria exhibit a range of ecological adaptations and interactions. While some are associated with human diseases, others are found in extreme environments and participate in sulfur metabolism and chemoautotrophy. Their diverse characteristics contribute to their ecological significance in various ecosystems.
Zeta proteobacteria: General characteristics with suitable examples
Zetaproteobacteria are a class of Gram-negative bacteria that are typically associated with marine Fe(II)-oxidizing environments. They exhibit several general characteristics and are involved in various ecological and biogeochemical processes. Here is an overview of their general characteristics along with suitable examples:
- General Characteristics:
- Gram-negative: Zetaproteobacteria have a cell wall structure that stains pink or red in the Gram stain, indicating the presence of an outer membrane.
- Chemolithoautotrophic: These bacteria obtain their energy by oxidizing inorganic compounds. Zetaproteobacteria specifically oxidize ferrous iron (Fe(II)) to ferric iron (Fe(III)).
- Microaerophilic: They thrive in environments with low oxygen levels and require oxygen as the terminal electron acceptor during Fe(II) oxidation.
- Morphological diversity: Zetaproteobacteria exhibit various morphotypes, including amorphous particulate oxides, twisted or helical stalks, sheaths, and Y-shaped irregular filaments.
- Examples of Zetaproteobacteria:
- Gallionella ferruginea: This bacterium is known to form helical stalks and is found in both freshwater and marine iron habitats.
- Sideroxydans lithotrophicus: It is a marine bacterium that participates in the oxidation of ferrous iron.
- Leptolyngbya sp.: Although primarily known as cyanobacteria, some species of Leptolyngbya have been observed to contribute to iron oxidation in freshwater iron-rich environments.
- Mariprofundus ferrooxidans: This bacterium is commonly found in marine environments and plays a crucial role in the biogeochemical cycling of iron.
- Zetaproteus sp.: This genus comprises various species of Zetaproteobacteria that contribute to the oxidation of ferrous iron.
Zetaproteobacteria are important in the biogeochemical cycling of iron, particularly in marine and freshwater environments. Their ability to oxidize ferrous iron to ferric iron helps facilitate the formation of iron oxides, which have significant implications for sedimentary rock formation and heavy metal binding. Additionally, these bacteria exhibit potential biotechnological applications, such as the production of enzymes and biominerals.
As research on Zetaproteobacteria continues, further discoveries and insights into their ecological roles and biotechnological potential are expected to emerge.
Importance of Gram Negative bacteria
Gram-negative bacteria play a significant role in various ecological, industrial, and medical contexts. Here are some key points highlighting the importance of Gram-negative bacteria:
- Ecological Balance: Gram-negative bacteria are crucial components of the ecological balance in diverse ecosystems. They contribute to nutrient cycling, decomposition, and the maintenance of ecosystem stability. These bacteria participate in processes such as nitrogen fixation, carbon cycling, and degradation of organic matter, playing essential roles in maintaining the health and functioning of ecosystems.
- Nutrient Cycling: Certain Gram-negative bacteria have the ability to fix atmospheric nitrogen into forms usable by other organisms. This nitrogen fixation is critical for the availability of nitrogen, an essential nutrient for the growth of plants and other organisms in ecosystems. Gram-negative bacteria, such as those in the family Rhizobiaceae, form mutualistic associations with leguminous plants, providing them with fixed nitrogen while obtaining nutrients from the plant.
- Symbiotic Relationships: Gram-negative bacteria establish symbiotic relationships with various organisms, including humans and animals. They inhabit the gastrointestinal tract, aiding in digestion, vitamin synthesis, and protecting against harmful pathogens. For example, Escherichia coli, a Gram-negative bacterium, produces vitamin K and helps prevent colonization by pathogenic bacteria.
- Bioremediation: Gram-negative bacteria have the ability to degrade and detoxify various environmental pollutants. They can break down harmful substances, such as hydrocarbons, pesticides, and heavy metals, through processes like biodegradation and bioremediation. This capability makes them valuable in cleaning up contaminated environments and reducing the impact of pollutants on ecosystems.
- Industrial Applications: Gram-negative bacteria are used in various industrial processes. They are employed in the production of enzymes, antibiotics, biofuels, and other valuable compounds through fermentation and biotechnological processes. Bacteria such as Escherichia coli and Pseudomonas putida have been genetically engineered to produce proteins, pharmaceuticals, and bio-based materials.
- Pathogenicity and Disease: While many Gram-negative bacteria are harmless or beneficial, some are responsible for causing diseases in humans, animals, and plants. Pathogens like Escherichia coli, Salmonella, Vibrio cholerae, and Pseudomonas aeruginosa can lead to severe infections and illnesses. Understanding the mechanisms of pathogenic Gram-negative bacteria is crucial for developing effective treatments and preventive measures.
- Antibiotic Resistance: Gram-negative bacteria are known for their ability to develop resistance to antibiotics. This poses a significant challenge in healthcare settings, as infections caused by resistant Gram-negative bacteria can be difficult to treat. Studying the resistance mechanisms and finding innovative solutions to combat antibiotic resistance in Gram-negative bacteria is of paramount importance in modern medicine.
In summary, Gram-negative bacteria play vital roles in ecological processes, nutrient cycling, symbiotic relationships, bioremediation, industrial applications, and disease. Understanding their biology, ecology, and pathogenicity is essential for harnessing their benefits while mitigating their negative impacts.
FAQ
What are Gram-negative bacteria?
Gram-negative bacteria are a group of bacteria characterized by their cell wall structure, which does not retain the violet crystal stain in the Gram staining method. Instead, they appear pink or red when counterstained.
What is the significance of Gram-negative bacteria?
Gram-negative bacteria are significant for several reasons. They are a common cause of various infectious diseases in humans and animals, including pneumonia, urinary tract infections, and gastrointestinal infections. Some Gram-negative bacteria also possess antibiotic resistance mechanisms, making them challenging to treat.
How do Gram-negative bacteria differ from Gram-positive bacteria?
Gram-negative bacteria have a more complex cell wall structure compared to Gram-positive bacteria. They have a thin peptidoglycan layer surrounded by an outer membrane composed of lipopolysaccharides (LPS), which provides them with additional protection and contributes to their pathogenic properties.
What are the common examples of Gram-negative bacteria?
Some common examples of Gram-negative bacteria include Escherichia coli, Salmonella spp., Klebsiella pneumoniae, Pseudomonas aeruginosa, Neisseria meningitidis, and Acinetobacter baumannii, among many others.
How do Gram-negative bacteria cause infection?
Gram-negative bacteria possess various virulence factors that allow them to invade and colonize host tissues. These include adhesive structures, toxins, and enzymes that help them evade the host immune response and cause tissue damage.
What are the antibiotic resistance mechanisms in Gram-negative bacteria?
Gram-negative bacteria are known for their ability to develop antibiotic resistance. They possess several mechanisms, such as efflux pumps that remove antibiotics from the bacterial cell, enzymes that inactivate antibiotics, and modifications in the outer membrane porins to limit antibiotic entry.
How are Gram-negative infections diagnosed?
Diagnosis of Gram-negative bacterial infections typically involves collecting clinical samples, such as blood, urine, or swabs from the affected site. These samples are then cultured and analyzed to identify the specific bacteria causing the infection.
How are Gram-negative infections treated?
Treatment of Gram-negative infections often involves the use of antibiotics effective against these bacteria. However, due to the rising antibiotic resistance, selecting appropriate antibiotics can be challenging. In some cases, combination therapy or alternative treatment options may be necessary.
Can Gram-negative bacteria be prevented?
Prevention of Gram-negative bacterial infections can be achieved through various measures, including practicing good hygiene, proper food handling and preparation, ensuring clean water sources, and implementing infection control practices in healthcare settings.
What research is being done on Gram-negative bacteria?
Research on Gram-negative bacteria focuses on understanding their mechanisms of pathogenesis, antibiotic resistance, and developing new treatment strategies. Additionally, studies are being conducted to develop vaccines targeting specific Gram-negative pathogens and to explore alternative antimicrobial approaches to combat these bacteria.
References
- Nikaido, H., & Nakae, T. (eds.). (2020). Bacterial Outer Membranes: Biogenesis and Functions. Springer.
- Silhavy, T. J., Kahne, D., & Walker, S. (2010). The bacterial cell envelope. Cold Spring Harbor Perspectives in Biology, 2(5), a000414.
- Nikaido, H. (2003). Molecular basis of bacterial outer membrane permeability revisited. Microbiology and Molecular Biology Reviews, 67(4), 593-656.
- Waksman, S. A. (1961). Classification, identification, and description of genera and species. In The Actinomycetes (Vol. 2, pp. 1257-1303). Springer.
- Tønjum, T. (2005). DNA transfer in the gram-negative bacteria. In The Horizontal Gene Pool (pp. 165-188). Springer.
- Nikaido, H. (2009). Multidrug resistance in bacteria. Annual Review of Biochemistry, 78, 119-146.
- Ventola, C. L. (2015). The antibiotic resistance crisis: part 1: causes and threats. Pharmacy and Therapeutics, 40(4), 277-283.
- CDC. (2019). Antibiotic Resistance Threats in the United States, 2019. Centers for Disease Control and Prevention.
- Lippa, A. M., Goulian, M., & Yan, J. (2014). Understanding how antibiotic resistance evolves: insights from the beta-lactamase TEM-1 and its recent variants. Evolutionary Applications, 7(7), 858-869.
- World Health Organization. (2017). Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. World Health Organization.


