Advertisements
Table of Contents
What are Ribozymes (RNA enzymes)?
- Ribozymes are catalytically active RNA molecules or RNA–protein complexes in which the RNA alone is responsible for the catalytic activity.
- The term ribozyme simultaneously refers to enzymatic activity and ribonucleic acid nature.
- Ribozymes are present in the genomes of all living kingdoms.
- Hairpin, hammerhead, Hepatitis delta virus (HDV), Varkud Satellite, and glmS ribozymes, which form the classes of small ribozymes, as well as group I and II introns, the ribosome, spliceosome, and RNase P, which are categorised as large ribozymes, are well-established natural ribozymes.
- Initially, RNA was considered only a messenger molecule between DNA, which stores genetic information, and proteins, which catalyse chemical events.
- In 1978, Sidney Altmann was the first to demonstrate that the RNA moiety was needed for catalysis in the RNA-cleaving RNA–protein complex RNase P from Escherichia coli.
- Later on, he established that the protein portion of RNase P was not required for catalysing the cleavage.
- Thomas Cech found in the early 1980s that the RNA of the protozoan Tetrahymena thermophila excises itself from the mRNA it dwells in and connects the two flanking ends.
- This self-splicing event was the first demonstration of an RNA’s catalytic activity in the absence of protein. In 1989, both Cech and Altmann were awarded the Nobel Prize in Chemistry.
Structure and mechanism of Ribozymes
- Ribozymes, like protein enzymes, have a specified three-dimensional structure for catalytic activity.
- Ribozymes require metal ions such as K+ or Mg2+, which are naturally present in the cell, in order to fold.
- These ions counterbalance the large density of negative charges within a folded oligonucleotide.
- The establishment of stable secondary structures typically precedes the formation of tertiary contacts.
- Large ribozymes can become caught in alternate conformations during folding, which must be undone before the catalytically active structure is achieved.
- In some instances, protein cofactors that aid in folding have been found.
- Except for the ribosome, all natural ribozymes catalyse phosphoryl transfer processes, which involve the nucleophilic substitution of one phosphodiester bond for another.
- Small ribozymes are categorised as 5′-OH displacement ribozymes, while group I and II introns, RNase P, and the spliceosome are 3′-OH displacement ribozymes.
- In tiny ribozymes, nucleophilic assault from the 2′-OH of the neighbouring upstream nucleotide cleaves the phosphodiester link.
- Large ribozymes create active sites that are intricate and employ an exogenous group for nucleophilic assault.
- These foreign groups can be water (RNase P, group II introns), an OH group of a mononucleotide or an internal nucleotide far from the cleavage site (group I and II introns, spliceosome), or the amine of the amino acid to be added to a developing peptide chain (ribosome).
- The two most prevalent processes employed by ribozymes are acid–base catalysis and direct metal ion-assisted catalysis, as seen in RNase P, the HDV ribozyme, and group I and II introns.
- In both methods, metal ions indirectly engage in catalysis by stabilising the transition state or the leaving group.
Splicing pathways of group I and II introns and the spliceosome
Group I
- In the initial step of group I intron splicing, the 3′-OH of an exogenous GTP binds to the backbone phosphate of the 5′-splice site, thereby releasing the 5′-exon.
- The 3′-OH of the 5′-exon attacks the 3′-splice site in the second phase.
- Breaking the link between the intron and the 3′-exon releases the two ligated exons and the intron in a linear form, with one guanosine residue added.
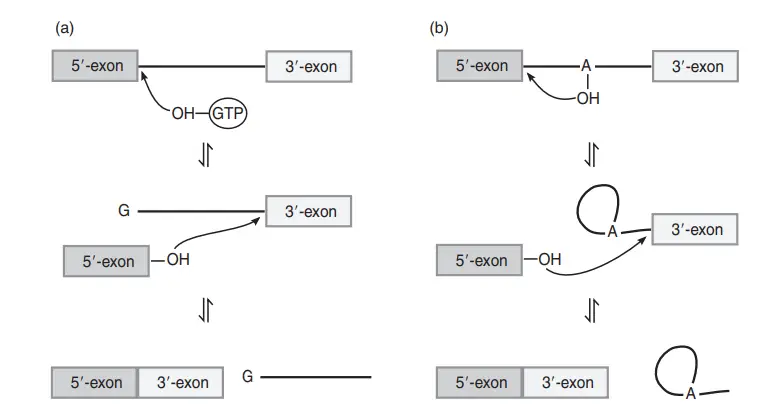
Group II
- In the initial step of group II intron and spliceosomal splicing, the 2′-OH of the intronic branch point adenosine attacks the 5′-splice site. The 5′-exon is released and an intron lariat structure is formed.
- The second step that links the exons proceeds similarly to group I introns, except the intron is released as a lariat. For group I and group II intron splicing, all stages are reversible.
Types of Ribozymes
There are the different types of ribozymes such as;
- RNase P Ribozyme
- Hammerhead ribozymes
- GIR1 branching ribozyme
- hepatitis delta virus (HDV) ribozyme hatchet ribozyme
- Pistol ribozyme
- VS ribozyme
- Twister ribozymes
- Twister sister ribozyme
- Group 1 introns
- Group2 introns
1. RNase P ribozyme
- RNase P is a crucial enzyme in tRNA production. It is present in all cells and organelles that synthesise tRNA.
- It is an RNA processing endonuclease that cleaves 5P-sequences and mature tRNAs from tRNA precursors.
- All known RNase P enzymes are ribonucleoproteins (RNPs) including an RNA catalytic subunit.
- An necessary RNA subunit and a protein subunit make up the enzyme. The protein component is necessary for in vivo and in vitro cleavage activity at physiological magnesium concentration.
- In the presence of high amounts of Mg++ ions, however, the RNA component can correctly cleave precursor tRNAs in vitro.
- An acceptor stem of the substrate recognises the ribozyme external guide sequence (EGS) during the interaction of the enzyme with its substrate.
- In addition, the 5’CCA3′ triplet at the 3′ end of the EGS is required for the cleavage reaction. Using a suitable EGS, this cleavage method is adaptable to any RNA.
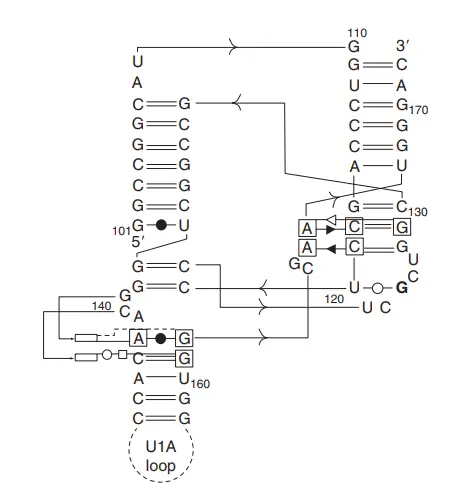
2. Hepatitis delta virus (HDV) ribozyme
- The hepatitis delta virus (HDV) ribozyme is a non-coding RNA required for viral replication, and it is the only human virus that uses ribozyme activity to infect its host.
- During replication of the hepatitis delta virus, which is believed to propagate by a double rolling circle mechanism, the ribozyme processes the RNA transcripts to unit lengths in a self-cleavage reaction.
- The ribozyme of the hepatitis delta virus is physically and biochemically similar to the ribozyme of the mammalian CPEB3 gene.
- Both positive and negative HDV strands are capable of self-cleaving in vitro to generate 5′ hydroxyl and 2′,3′-cyclic phosphate products.
- The analysis of the self-cleavage sites at nucleotide positions 658/686 (positive strand) and 900/901 (negative strand) revealed that neither of these regions resembles those of hammerhead or hairpin ribozymes.
- Two types of secondary structures have been proposed thus far: a pseudo-knot structure and an axe-head structure.
- The structure resembling a pseudo-knot is formed of four stems labelled I, II, III, and IV, respectively.
- The first two stems form the pseudo-knot-like shape, whereas the third and fourth stems adopt more typical stem and loop topologies, respectively.
- The axe-head model predicts three stems, with the cleavage between stems III and I.
- Both models contain substrate and ribozyme within the same molecule. It is feasible to remove the ribozyme from its substrate, though.
- As the maintenance of a substrate’s well-defined secondary structure is necessary for the activity of HDV ribozymes, it is challenging to identify excellent substrates that are not natural.
3. Hammerhead Ribozyme
- At a precise location within an RNA molecule, the hammerhead ribozyme catalyses reversible cleavage and ligation events.
- It is one of numerous naturally occurring catalytic RNAs (ribozymes). The hammerhead ribozyme is structurally constructed of three base-paired helices that are separated by short linkers of conserved sequences. These helices are designated as I, II, and III.
- The 5′ and 3′ ends of hammerhead ribozymes can be categorised into three categories based on the helix in which they are located.
- If the 5′ and 3′ ends of the sequence contribute to stem I, it is a type I hammerhead ribozyme. If they contribute to stem II, it is a type II, and if they contribute to stem III, it is a type III.
- Of the three conceivable topological kinds, type I is found in the genomes of prokaryotes, eukaryotes, and RNA plant pathogens, whereas types II and III are predominantly found in plants, plant pathogens, and prokaryotes, respectively.
- The hammerhead ribozymes, whose name derives from their secondary structure, are cis-acting elements in their natural context, meaning that the ribozyme and its substrate are part of the same molecule.
- It was established, however, that the hammerhead RNA from the (+) strand of TobRV (tobacco ringspot virus) could be separated into a substrate and an enzyme bearing the catalytic activity.
- Thus, these ribozymes can perform a real enzymatic process in which a substrate is cleaved without altering the ribozyme itself.
- The currently recognised structural model for trans-acting hammerhead ribozymes has three crucial characteristics:
- Three double helices labelled I, II, and III;
- a cleavage site on the target RNA composed of the trinucleotide triplet GUC in the majority of naturally occurring ribozymes;
- two lengths of highly conserved sequence 5’CUGAXGA and 5’GAAA, where X is any base except G.
- Natural satellite and viroid RNAs, with the exception of one, cleave at a GUC target site. Other triplets, however, can be sliced with comparable or even greater efficiency.
- In every instance, cleavage happens at the 3′ end of the final base of the triplet.
- The substrate molecule’s 5′-hydroxyl and 2′-3′-cyclic phosphate termini are severed by a Mg++-dependent transesterification reaction catalysed by hammerhead ribozymes.
- The two lengths of conserved nucleotides (5’CUGAXGA and 5’GAAA) are crucial to the cleavage step, as their substitution drastically reduces ribozyme activity efficiency.
- Due to the fact that helices I and III are not conserved, it is possible to create ribozymes with flanking sections that are homologous to any substrate sequence.
- Nonetheless, the length and sequencing of surrounding regions can influence the efficacy of cleavage.
- Several investigations on kinetics have demonstrated that hammerhead ribozymes with short flanking regions function as genuine catalysts in vifro.
- A simple kinetic description of one turnover reaction includes I assembly of ribozyme and substrate into a ribozyme-substrate complex, (ii) breaking of phosphodiester bond, resulting in the formation of two products that remain bound to the ribozyme, and (iii) release of the product.
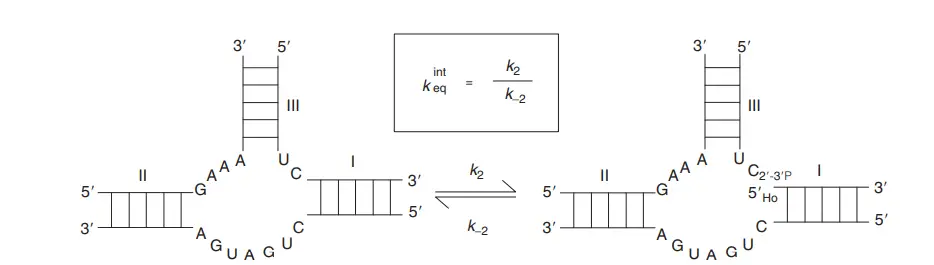
defined by the constant of cleavage k2 and the reaction of ligation is defined by the constant of ligation k–2. The definition of the
equilibrium constant is framed.
4. Hairpin Ribozyme
- The hairpin ribozyme is a short RNA segment that can function as a ribozyme.
- Similarly to the hammerhead ribozyme, it is present in the RNA satellites of plant viruses.
- It was initially found in the minus strand of the tobacco ringspot virus (TRSV) satellite RNA, where it catalyses self-cleavage and joining (ligation) events to transform rolling circle virus replication products into linear and circular satellite RNA molecules.
- Similar to the hammerhead ribozyme, the hairpin ribozyme does not require a metal ion for the reaction.
- The hairpin ribozyme is an RNA motif that catalyses RNA processing events necessary for the replication of satellite RNA molecules in which it is contained.
- These are self-processing reactions, in which a molecule rearranges its own structure. The ribozyme motif mediates both cleavage and end joining processes, resulting in a combination of interconvertible linear and circular satellite RNA molecules.
- These processes are essential for the processing of the massive multimeric RNA molecules produced by rolling circle replication.
- At the conclusion of the replication cycle, these enormous intermediates of satellite RNA replication are reduced to unit-length molecules (circular or linear) prior to being packed by viruses and sent to neighbouring cells for additional rounds of replication.
5. Group I introns
- Group I intron splicing involves two consecutive ester-transfer events.
- The exogenous guanosine or guanosine nucleotide (exoG) initially docks onto the active G-binding site in P7, and its 3-OH is aligned to attack the phosphodiester bond at the 5 splice site in P1, resulting in a free 3′-OH group at the upstream exon and the exoG being attached to the 5′ end of the intron.
- The terminal G (omega G) of the intron then replaces the exoG and occupies the G-binding site to organise the second ester-transfer reaction: the 3′-OH group of the upstream exon in Pl is aligned to attack the 3′ splice site in P10, resulting in the ligation of the adjacent upstream and downstream exons and the release of the catalytic intron.
- It was hypothesised that group I and group II introns use the two-metal-ion mechanism seen in protein polymerases and phosphatases to perform phosphoryl transfer processes, (Which was unequivocally shown by a newly determined high-resolution structure of the Azoarcus group I intron.
6. Group II introns
- Group II introns are a huge family of self-catalytic ribozymes and mobile genetic elements present in all three domains of life.
- Ribozyme activity (e.g., self-splicing) can occur in vitro in conditions of high salt concentration.
- However, protein assistance is required for in vivo splicing. In contrast to group I introns, intron excision happens in the absence of GTP and entails the creation of a lariat with an A-residue branchpoint that resembles lariats created during nuclear pre-mRNA splicing.
- Due to the same catalytic mechanism and structural similarities between the Domain V substructure and the U6/U2 extended snRNA, it is suggested that pre-mRNA splicing may have developed from group II introns.
7. Group -I- like ribozymes (GIR1) branching ribozyme
- The Lariat capping ribozyme (formerly known as the GIR1 branching ribozyme) is a 179-nucleotide ribozyme that resembles a group I ribozyme.
- It is located within a form of group I introns known as twin-ribozyme introns.
- Instead of splicing, it catalyses a branching process involving a nucleophilic assault on a neighbouring phosphodiester bond by the 2’OH of an internal residue.
- Consequently, the RNA is cleaved at an internal processing site (IPS), leaving behind a 3’OH and a downstream product with a small lariat at its 5′ terminus.
- The lariat is referred to as the lariat cap’ because it caps an intron-encoded mRNA.
- The resultant lariat cap appears to increase the half-life of Homing Endonuclease mRNA.
8. Glucosamine-6-phosphate riboswitch ribozyme (gim ribozyme)
- glm S ribozyme is an RNA structure that sits in the 5′ untranslated region (UTR) of the mRNA transcript of the gim gene.
- Upon activation, this RNA regulates the gim gene by responding to the quantity of a particular metabolite, glucosamine-6 phosphate (GlN6P), and activating a self-cleaving chemical process.
- This cleavage causes the degradation of the mRNA containing the ribozyme and reduces GlcN6P synthesis.
- The glms gene genes for glutamine-fructose-6-phosphate amidotransferase, an enzyme that catalyses the creation of GlcN6P, an important component for cell wall biosynthesis, from fructose-6-phosphate and glutamine.
- In the absence of GIN6P, the gene continues to be translated into glutamine-fructose-6-phosphate amidotransferase and GlcN6P is generated.
- GlcN6P is a cofactor for this cleavage reaction because it acts as an acid-base catalyst directly.
- This RNA is the first riboswitch known to be a self-cleaving ribozyme; it was identified, like many others, using bioinformatics.
9. Hatchet ribozyme
- The hatchet ribozyme is a structure made of RNA that cuts itself at a certain site. In other words, it is a ribozyme that can cut itself.
- A method called RNAs Associated with Genes Associated with Twister and Hammerhead ribozymes, or RAGATH, was used to find Hatchet ribozymes.
- Later biochemical analysis supports the idea that a ribozyme has a function and gives more details about the chemical reaction that the ribozyme speeds up.
10. Pistol ribozyme
- The pistol ribozyme is a structure made of RNA that cuts itself at a certain site.
- Internal phosphoester transfer is cut by pistol ribozymes.
11. VS ribozyme
- A phosphodiester bond is broken by the Varkud satellite (VS) ribozyme, which is an RNA enzyme.
- The largest known nucleolytic ribozyme, called Varkud satellite (VS)ribozyme, was found to be a part of VS RNA.
- VS RNA is a long non-coding RNA that is found as a satellite RNA in the mitochondria of Neurospora strains Varkud-1C and a few others.
- There are parts of both catalytic RNAs and group 1 introns in VS ribozyme. VS ribozyme has both cleavage and ligation activity, which means that it can do both cleavage and ligation reactions well even without proteins.
- VS ribozymes share genes with other Neurospora strains through horizontal gene transfer.
- VS ribozymes are different from other nucleolytic ribozymes in every way.
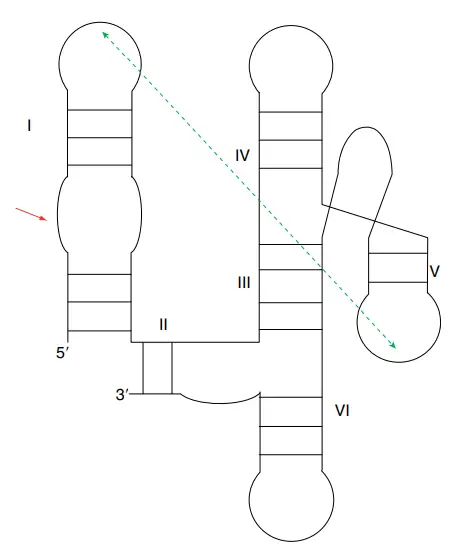
- VS RNA has a unique structure at the first, second, and third levels.
- The VS ribozyme has a secondary structure that is made up of six helical domains.
- The substrate domain is made up of stem loop I, and the catalytic domain is made up of stem loops II–VI. When these two domains are made separately in vitro, they can work together to do the self-cleavage reaction.
- Two helices make a cleft in which the substrate fits. A756, a very important nucleotide, is likely to be the site where the ribozyme does its work.
- The A730 loop and the A756 nucleotide are very important to how it works because they are involved in the phosphoric transfer chemistry that the ribozyme does.
12. Twister ribozyme
- The twister ribozyme is an RNA structure that can break itself apart.
- This rbozyme has been shown to have nucleolytic activity both in vivo and in vitro, and it has one of the fastest catalytic rates of naturally occurring ribozymes with the same function.
- The twister ribozyme is thought to be a member of the small self-cleaving ribozyme family, which also includes the hammerhead, hairpin, hepatitis delta virus (HDV), Varkud satellite (VS), and glmS ribozymes.
- Like other nucleolytic ribozymes, the twister ribozyme breaks phosphodiester bonds selectively into a 2′,3′-cyclic phosphate and a 5′ hydroxyl. This is similar to how other nucleolytic ribozymes work.
13. Twister sister ribozyme
- The twister sister ribozyme (TS) is a structure made of RNA that cuts itself in a certain place. In other words, it is a ribozyme that can cut itself.
- A bioinformatics method called RAGATH, which stands for RNA Associated with Genes Associated with Twister and Hammerhead ribozymes, was used to find the twister sister ribozyme.
- The structure of the twister sister ribozyme might be similar to that of the twister ribozyme. Most of the time, nucleolytic ribozymes use the SN2 mechanism to break a specific phosphodiester linkage.
- The O2′ acts as a nucleophile to attack the nearby P, and the 05′ is a leaving group. The products of catalysis are a cyclic 2,3′ phosphate and a hydroxyl at the 5′ end.
- The activity of twister sister as a catalyst goes up as the pH goes up and depends on the type of metal ion.
- The rate of cleavage doubles for every unit increase in pH and reaches a plateau near pH 7.
14. Artificial Ribozymes
- Since ribozymes were found in living things, people have been interested in learning more about ribozymes that are made in the lab. For example, RNAs that can cut themselves and have good enzyme activity have been made in a lab. Tang and Breaker found self-cleaving RNAs by picking out RNAs from random-sequence RNAs in the lab. Some of the synthetic ribozymes made had new structures, while others were like the hammerhead ribozyme, which is found in nature.
- Darwinian evolution is used in the methods used to find artificial ribozymes. This method takes advantage of the fact that RNA is both a catalyst and a polymer that stores information. This makes it easy for researchers to make large numbers of RNA catalysts using polymerase enzymes. The ribozymes are changed by using reverse transcriptase to turn them into different types of cDNA, which are then amplified with mutagenic PCR. In these experiments, the selection criteria are often different. Using biotin tags that are chemically linked to the substrate is one way to choose a ligase ribozyme. A streptavidin matrix can be used to find the active molecules in a molecule that has the desired ligase activity.
- c. Lincoln and Joyce made a system of RNA enzymes that can copy itself in about an hour. Using in vitro evolution on a mixture of candidate enzymes led to the formation of a pair of RNA enzymes that can make each other from synthetic oligonucleotides without any protein.
Mechanisms of ribozyme catalysis
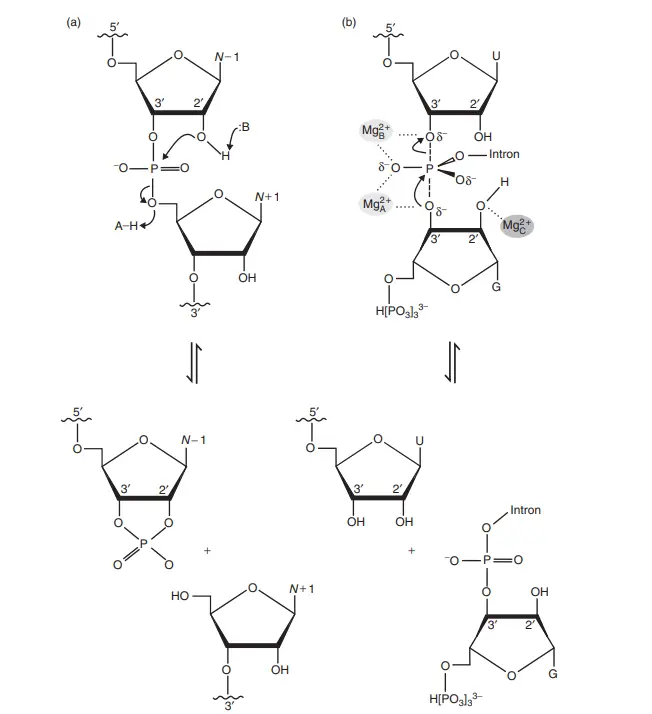
- Acid–base catalysis as observed in the hairpin ribozyme: B is a generic base that deprotonates the 3′-OH group in order to activate it as a nucleophile. A–H is a generic acid that stabilises the leaving group through proton donation. The 2′-OH assault on the neighbouring phosphodiester results in the formation of a 2′,3′-cyclic phosphate and a 5′-hydroxyl terminal.
- Metal ion-assisted catalysis: On the basis of the first stage of group I intron splicing, a typical two-metal-ion mechanism is illustrated. In this instance, every metal ion is Mg2+. The two metal ions that participate directly in catalysis are Mg2+ A, which can bind OH ions in solution and accept the proton from the OH nucleophile as a general base, and Mg2+ B, which accepts electrons from the 3′-oxyanion of the intron and stabilises it as a leaving group. Mg2+ C further stabilises the transition state by coordination of the 2′-OH of GTP. In the case of RNase P, strand cleavage produces a free 3′-hydroxyl group and a new phosphodiester bridge (group I and II introns, spliceosome) or a free phosphate.
Applications of Ribozymes
- Catalytic RNAs, also called ribozymes, can cut RNA molecules in a specific way. This makes them possible antiviral and anticancer drugs, as well as powerful tools for studying how genomes work.
- In recent years, ribozymes have been used successfully to stop the expression of genes in both in vitro and in vivo (living) biological systems.
- Ribozyme gene therapy has been used to treat AIDS patients in Phase I clinical trials.
- Gene shears, a type of synthetic ribozyme that cuts apart HIV RNA, has been made and is now being tested in people with HIV.
References
- Symons, R. H. (1999). RIBOZYMES. Encyclopedia of Virology, 1551–1559. doi:10.1006/rwvi.1999.0254
- Skilandat, M. & Paulus, Susann & Sigel, R.K.O.. (2017). Ribozymes. 10.1016/B978-0-12-809633-8.07077-1.
- Westhof, E., & Lescoute, A. (2008). Ribozymes. Encyclopedia of Virology, 475–481. doi:10.1016/b978-012374410-4.00495-7
- FEMS Microbiology Reviews, Volume 23, Issue 3, June 1999, Pages 257–275, https://doi.org/10.1111/j.1574-6976.1999.tb00399.x
- Grassi G, Marini JC. Ribozymes: structure, function, and potential therapy for dominant genetic disorders. Ann Med. 1996 Dec;28(6):499-510. doi: 10.3109/07853899608999114. PMID: 9017109.
- https://www.news-medical.net/life-sciences/What-are-Ribozymes.aspx
- https://www.wiley.com/en-us/Ribozymes:+Principles,+Methods,+Applications,+2+Volume+Set-p-9783527344543
- https://www.bionity.com/en/encyclopedia/Ribozyme.html
- https://www2.chem.wisc.edu/deptfiles/genchem/netorial/modules/biomolecules/modules/enzymes/enzyme9.htm
- http://scottlab.ucsc.edu/scottlab/reprints/2007_Scott_Curr_Op_Str_Biol.pdf
- https://www.slideshare.net/swormage/ribozyme-172078511


