Table of Contents
What is Ribosomal RNA (rRNA)?
- Ribonucleic acid refers to the RNA contained in ribosomes, the molecules responsible for catalysing protein synthesis (rRNA).
- Over 60-80% of the ribosome’s mass is formed of ribosomal RNA, which is important for all of the ribosome’s actions, such as binding to messenger RNA, attracting transfer RNA, and facilitating the synthesis of peptide bonds between amino acids.
- A ribosome’s structure is influenced by the three-dimensional structure of a rRNA core.
- By interacting with the core, ribosomal proteins contribute to the maintenance of this structure.
- The translation of ribosomal RNA takes place within the nucleus’ nucleoli. These are dense, spherical formations that form around rRNA-coding genes.
- The eventual creation of ribosomes is contingent on the retention of ribosomal proteins by nucleoli.
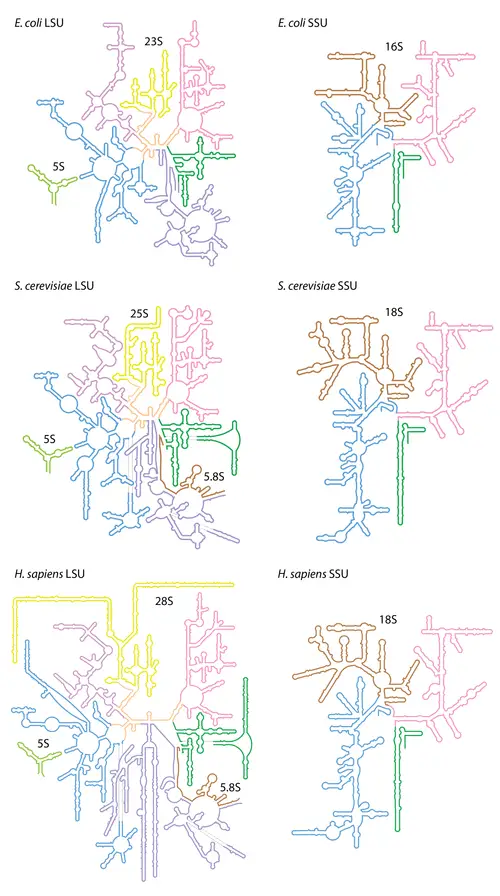
- In prokaryotic cells, there are three unique sizes of rRNA (23S, 16S, and 5S). There are four rRNA species in the eukaryotic cytosol (28S, 18S, 5.8S, and 5S, where “S” is the Svedberg unit for sedimentation rate, which is regulated by the size and shape of the particle).
Discovery of Ribosomal RNA
- During cell fractionation research studying the role of RNA viruses in developing cancer, ribosomal RNA was identified.
- Fractionation is a technique in which cell membranes are removed carefully and selectively, while maintaining the function of cellular organelles. The homogenised cytoplasm is then centrifuged at varying speeds to separate the organelles by density.
- Initial investigations that discovered the existence of rRNA extracted a fraction assumed to represent a novel subcellular organelle specialised in protein synthesis called the microsome.
- Later, it was determined that ribosomes on the endoplasmic reticulum were responsible for the detection of RNA in these samples.
- Since ribosomal subunits and rRNA were initially found by differential centrifugation, they are still distinguished by their sedimentation rate, as measured by the Svedberg coefficient.
- However, as they are not molecular weight measurements, the coefficients cannot be added directly. For instance, when the 50S bigger subunit and 30S smaller subunit combine in bacterial ribosomes, the combination has a Svedberg coefficient of 70S.
Structure of rRNA
- Despite the fact that the fundamental structure of rRNA sequences might differ between organisms, base-pairing within these sequences typically produces stem-loop arrangements.
- The length and position of these rRNA stem-loops allow them to form species-specific three-dimensional rRNA structures.
- rRNA can develop tight and specialised connections with ribosomal proteins to generate ribosomal subunits due to various configurations.
- These ribosomal proteins have basic residues (as opposed to acidic residues) and aromatic residues (i.e. phenylalanine, tyrosine, and tryptophan) that enable them to develop chemical interactions with their corresponding RNA sections, including stacking interactions.
- Ribosomal proteins can cross-link to the sugar-phosphate backbone of rRNA via basic residue-containing binding sites (i.e. lysine and arginine).
- All ribosomal proteins have been identified, including the particular sequences that bind to rRNA. Together with the attachment of the small and large ribosomal subunits, these interactions produce a functional ribosome capable of protein synthesis.
- Ribosomal RNA forms two primary ribosomal subunits: the large subunit (LSU) and the small subunit (SSU) (SSU).
- One of each type is required for the formation of a functional ribosome. Occasionally, the subunits are identified by their size-sedimentation measures (a number with an “S” suffix).
- In prokaryotes, the LSU and SSU are designated by the designations 50S and 30S, respectively. The LSU and SSU of eukaryotes are designated as the 60S and 40S subunits, respectively.
- In the ribosomes of prokaryotes such as bacteria, the SSU includes one short rRNA molecule (1500 nucleotides) and one large rRNA molecule (3000 nucleotides).
- These are joined with 50 ribosomal proteins to create subunits of ribosomes. Prokaryotic ribosomes have three forms of rRNA: 23S and 5S rRNA in the LSU and 16S rRNA in the SSU.
- In the ribosomes of eukaryotes such as humans, the SSU contains a single tiny rRNA (1800 nucleotides) and the LSU includes two small rRNAs and one giant rRNA molecule (5000 nucleotides).
- In compared to prokaryotes, eukaryotic rRNA contains over 70 ribosomal proteins that interact to produce larger and more polymorphic ribosomal units.
- In eukaryotes, there are four kinds of rRNA: three in the LSU and one in the SSU.
- Yeast has been the usual model for observing the behaviour and operations of eukaryotic rRNA, resulting in a lack of study diversity.
- Technical advancements (particularly in the field of Cryo-EM) have only enabled basic research into ribosomal behaviour in other eukaryotes over the last decade.
- The LSU contains the 5S, 5.8S, and 28S rRNAs in yeast. The combined 5.8S and 28S are broadly comparable in size and function to the bacterial 23S rRNA subtype, except the expansion segments (ESs) that are confined to the surface of the ribosome and were previously believed to be exclusive to eukaryotes.
- The Asgard phyla Lokiarchaeota and Heimdallarchaeota, the closest archaeal relatives to Eukarya, were reported to have two supersized ESs in their 23S rRNAs very recently.
- Likewise, the 5S rRNA of the ribosomes of the halophilic archaeon Halococcus morrhuae contains an insertion of 108 nucleotides.
- A eukaryotic SSU contains the portion of 18S rRNA that also contains ESs. In general, SSU ESs are smaller than LSU ESs.
- SSU and LSU rRNA sequences are commonly employed in the investigation of evolutionary relationships between organisms due to their antiquity, universal distribution, and resistance to horizontal gene transfer.
- Due to their essential role in ribosome activity, rRNA sequences remain conserved (not altered) over time.
- By estimating nucleotide similarity, phylogenetic information generated from the 16s rRNA is now the primary method for distinguishing between closely related bacterial species. The lineage of the translation system is the canonical tree of life.
- LSU rRNA subtypes are known as ribozymes because ribosomal proteins cannot attach to the ribosome’s catalytic site in this region (specifically the peptidyl transferase center, or PTC).
- The SSU rRNA subtypes decode messenger RNA at its decoding centre (DC). Ribosomal proteins are inadmissible to the DC.
- During the translation of other mRNAs, the structure of rRNA can dramatically alter to influence tRNA interaction with the ribosome.
- This is believed to occur in 16s rRNA when particular nucleotides appear to alternate base pairing between one nucleotide and another, generating a “switch” that affects the shape of the rRNA.
- This mechanism can modify the shape of the LSU and SSU, indicating that this conformational switch in the rRNA structure influences the entire ribosome’s capacity to match a codon with its anticodon in tRNA selection and decode mRNA.
Detail Structure of rRNA
5S ribosomal RNA
- 5S ribosomal RNA (5S rRNA) is a ribosomal RNA molecule with a length of around 120 nucleotides and a mass of 40 kDa.
- Except for the mitochondrial ribosomes of fungi and mammals, it is a structural and functional component of the major subunit of the ribosome in all domains of life (bacteria, archaea, eukaryotes).
- 5S refers to the sedimentation velocity of the molecule in an ultracentrifuge, which is measured in Svedberg units (S).
- In prokaryotes, the 5S rRNA gene is normally situated downstream of the small and large subunit rRNA genes in the rRNA operons and is co-transcribed into a polycistronic precursor.
- The presence of numerous 5S rRNA gene copies (5S rDNA) grouped in tandem repeats is unique to eukaryotic nuclear genomes, with the number of copies varies among species.
- Other eukaryotic rRNAs are cleaved from a 45S precursor transcribed by RNA polymerase I, but 5S rRNA is produced by RNA polymerase III.
- It has been demonstrated that fingers 4–7 of the nine-zinc finger transcription factor TFIIIA can bind to the core region of 5S RNA in Xenopus oocytes.
- The interaction between 5S rRNA and TFIIIA inhibits the transcription of the 5S RNA gene and stabilises the 5S RNA transcript until it is necessary for ribosome assembly.
5S ribosomal RNA Structure
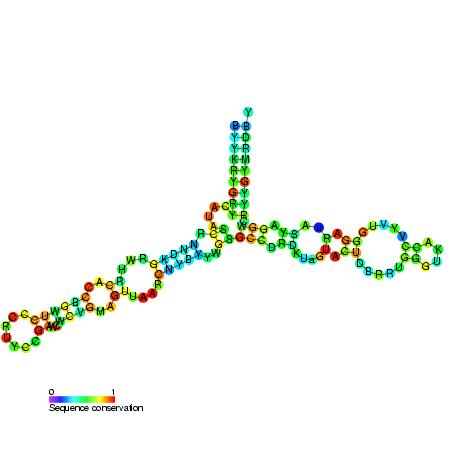
- Five helices (I-V in roman numerals), four loops (B-E), and one hinge (A) comprise the Y-shaped secondary structure of 5S rRNA. Loops B and E are internal hairpins, while loops C and D are terminal hairpins.
- Phylogenetic analyses indicate that helices I and III are likely ancestral.
- Helix III has two adenosines that are well conserved.
- With its hairpin configuration, Helix V is believed to interact with TFIIIA.
Location within the ribosome
- 5S rRNA is contained within the big component of the ribosome.
- In bacteria and archaea, the large ribosomal subunit (LSU) is composed of two RNA moieties, 5S rRNA and 23S rRNA, as well as other related proteins.
- The LSU of eukaryotes contains 5S, 5.8S, and 28S rRNAs as well as other proteins.
Functions of 5s rRNA
- tRNAs are bound by 5S rRNA-binding proteins and other proteins of the central protuberance of the LSU.
- 5S rRNA acts as a mediator between the two functional centres of the ribosome by establishing intersubunit bridges and tRNA-binding sites with 5S rRNA-binding proteins and other components of the central protuberance.
23S ribosomal RNA Structure
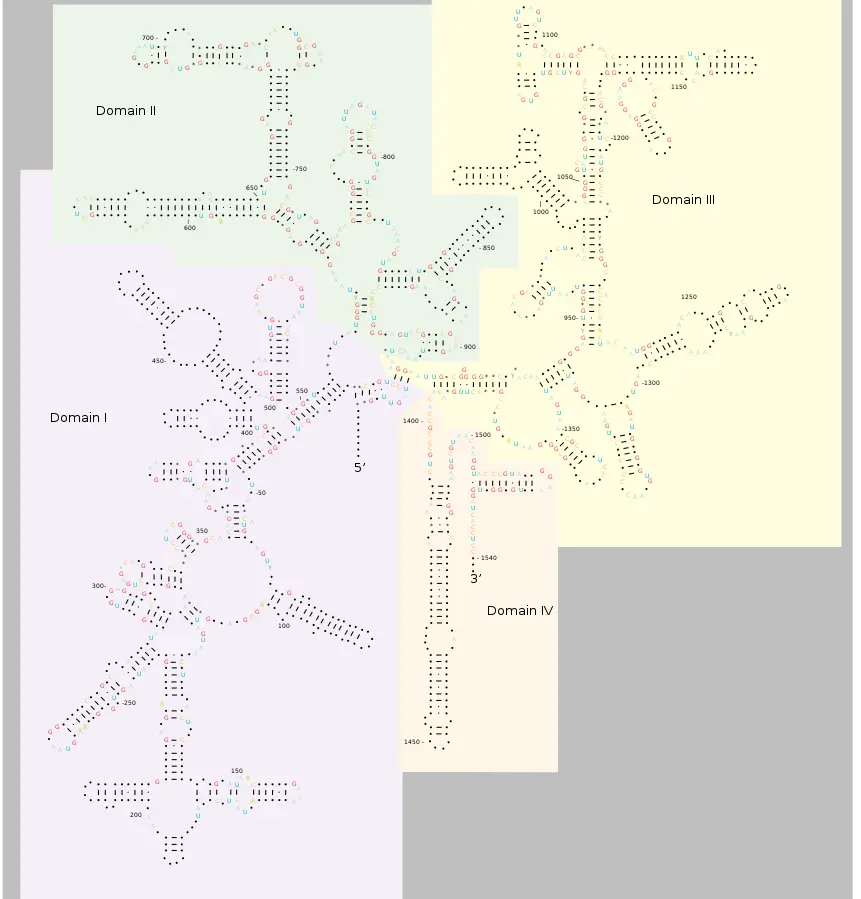
- The 23S rRNA is a 2,904-nucleotide-long (in E. coli) component of the 50S major subunit of the bacterial/archean ribosome that forms the peptidyl transferase core (PTC).
- The 23S rRNA is composed of six secondary structural domains labelled I-VI, whereas the 5S rRNA corresponds to domain VII.
- Domain V of this rRNA contains the ribosomal peptidyl transferase activity. Domain V is also the most common binding site for antibiotics that block translation, making it a target for ribosomal engineering.
- Chloramphenicol, a well-known member of this antibiotic class, inhibits the formation of peptide bonds, with new 3D structural investigations revealing two distinct binding sites depending on the ribosome species.
- Numerous mutations in Peptidyl transferase-active regions of 23S rRNA have led to antibiotic resistance.
- Typically, 23S rRNA genes have more sequence variants, including insertions and/or deletions, than other rRNA genes.
- The 28S ribosomal RNA, with a portion filled by the 5.8S ribosomal RNA, is the eukaryotic homolog of the 23S LSU rRNA.
23S rRNA Functions
- Peptidyl transferase is a key function of rRNA in general. The stimulating core of the ribosome is involved in the configuration of peptide bonds. Important for protein synthesis and transpeptidation response are both peptidyl-tRNA and aminoacyl-tRNA.
16S ribosomal RNA Structure
- 16S ribosomal RNA (16S rRNA) is the RNA component of a bacterial ribosome’s 30S subunit (SSU rRNA).
- It provides the majority of the SSU structure and binds to the Shine-Dalgarno sequence.
- Due to the slow rates of evolution of this region of the gene, the 16S rRNA gene is utilised in reconstructing phylogenies.
- In 1977, Carl Woese and George E. Fox were two of the first people to employ 16S rRNA in phylogenetics.
- Multiple 16S rRNA gene sequences can exist inside a same bacterium.
Functions of 16S ribosomal RNA
- As with the big (23S) ribosomal RNA, it has a structural function, establishing the locations of the ribosomal proteins as a scaffold.
- The 3′-end includes the anti-Shine-Dalgarno region, which binds upstream of the mRNA’s AUG start codon. The 3′-end of 16S RNA interacts to the proteins S1 and S21 that have been implicated in the start of protein synthesis.
- Interacts with 23S to facilitate the association of the two ribosomal subunits (50S and 30S)
- The formation of a hydrogen bond between the N1 atoms of adenine residues 1492 and 1493 and the 2′OH group of the mRNA backbone stabilises proper codon-anticodon pairing at the A-site.
28S ribosomal RNA Structure
- As the structural ribosomal RNA (rRNA) for the large subunit (LSU) of eukaryotic cytoplasmic ribosomes, 28S ribosomal RNA is a fundamental component of all eukaryotic cells. It is 25S in plants and 28S in mammals, hence the alternative name 25S–28S rRNA.
- It is the eukaryotic nuclear homologue of the bacterial 23S and mitochondrial 16S ribosomal RNAs when combined with the 5.8S rRNA on the 5′ end.
- The average length of the 28S rRNA is between 4000 and 5000 nucleotides.
- Before assembling 28S rRNA into the ribosome, certain eukaryotes cleave it into two pieces, a process known as the “hidden break.”
Functions
- The genes that code for 28S rRNA are known as 28S rDNA. Comparing the sequences of these genes is occasionally employed in molecular analysis to create phylogenetic trees, such as in protists, fungi, insects, arachnids, tardigrades, and vertebrates.
18S ribosomal RNA Structure
- 18S ribosomal RNA (18S rRNA) is a component of the ribosomal RNA.
- The 18S stands for Svedberg units. 18S rRNA is an SSU rRNA, a component of the ribosomal small subunit in eukaryotes (40S).
- 18S rRNA is the structural RNA for the small ribosomal subunit of eukaryotic cytoplasmic ribosomes, and is therefore one of the fundamental components of all eukaryotic cells.
- 18S rRNA is the eukaryotic cytosolic homologue of prokaryotic and plastid 16S ribosomal RNA. In mitochondria, 18S rRNA is a homolog of 12S ribosomal RNA.
- 18S rRNA-encoding genes are referred to as 18S rRNA genes. Sequence information from these genes is commonly employed in molecular analysis to reconstruct the evolutionary history of animals, particularly vertebrates, because to their slow evolutionary rate, which makes it excellent for reconstructing ancient divergences.
Functions
- The small subunit (SSU) 18S rRNA gene is one of the most commonly employed genes in phylogenetic research and an essential marker for random target polymerase chain reaction (PCR) in environmental biodiversity screening.
5.8S ribosomal RNA Structure
- 5.8S ribosomal RNA (5.8S rRNA) is a non-coding RNA component of the major subunit of the eukaryotic ribosome and hence plays a crucial role in protein translation.
- RNA polymerase I transcribes it as part of the 45S precursor, which also comprises 18S and 28S rRNA. It is believed to facilitate ribosome translocation.
- Additionally, it is known to create covalent bonds with the tumour suppressor protein p53.
- 5.8S rRNA can serve as a reference gene for the identification of microRNAs. The 5.8S ribosomal RNA is utilised to gain a deeper understanding of various rRNA processes and pathways within the cell.
- The 5′ end of the non-eukaryotic LSU rRNA is homologous to the 5′ end of the 5.8S rRNA. The inclusion of ITS2 in eukaryotes separates LSU rRNA into 5.8S and 28S rRNAs. In other insects, the 5.8 rRNA is further divided into two pieces.
Types of rRNA
In Prokaryotes
- The 16S ribosomal RNA in prokaryotes is contained within a compact 30S ribosomal subunit.
- Two rRNA species are found in the 50S subunit of the big ribosome (the 23S and 5S ribosomal RNAs).
- Therefore, it may be argued that in archaea and bacteria, a single rRNA gene codes for the three rRNA types: 16S, 23S, and 5S.
- The 23S, 16S, and 5S rRNA genes of bacteria are often organised as a co-transcribed operon.
In Eukaryotes
- The eukaryotic ribosome consists of a 40S subunit and a 60S subunit. In contrast, eukaryotes typically have many versions of the rRNA genes organised in repetitive sequences.
- There are around 300-400 repeats in five clusters on human chromosomes: 13 (RNR1), 14 (RNR2), 15 (RNR3), 21 (RNR4) and 22 (RNR5) (RNR5).
- rRNA is also present in chloroplasts and mitochondria of eukaryotic cells. Ribosomes can be found as free-floating complexes or attached to the endoplasmic reticulum in the cytoplasm.
Degradation of rRNA
- Compared to other common kinds of RNA, ribosomal RNA is quite stable and remains for longer durations in a healthy cellular environment.
- Once formed into functional units, ribosomal RNA is stable in the stationary phase of the cell’s life cycle for several hours.
- Stalling of a ribosome, which occurs when the ribosome detects incorrect mRNA or faces other processing challenges that halt translation by the ribosome, can initiate degradation.
- Once a ribosome becomes stalled, a specific route on the ribosome is activated to target the entire complex for breakdown.
In eukaryotes
As with the creation of any protein or RNA, rRNA production is susceptible to mistakes that result in the production of nonfunctional rRNA. To address this, the cell permits the nonfunctional rRNA decay (NRD) pathway to degrade rRNA. The majority of this topic’s study has been undertaken on eukaryotic cells, notably Saccharomyces cerevisiae yeast. How eukaryotic cells are able to target functionally faulty ribosomes for ubiquitination and destruction is now only partially understood.
- It is possible that the NRD pathway for the 40S subunit is independent or distinct from the NRD pathway for the 60S subunit. Certain genes have been discovered to influence the degradation of particular pre-RNAs, but not others.
- Numerous proteins, such as Mms1p and Rtt101p, are hypothesised to bind together to target ribosomes for degradation as part of the NRD pathway. Rtt101p is expected to recruit a ubiquitin E3 ligase complex, allowing non-functional ribosomes to be ubiquitinated prior to degradation.
- It is unknown how prokaryotes are able to destroy non-functional rRNAs given that they lack a Mms1 homolog.
- The accumulation of nonfunctional rRNAs did not appear to significantly impair the growth rate of eukaryotic cells.
In prokaryotes
Despite the fact that there is significantly less research on ribosomal RNA degradation in prokaryotes compared to eukaryotes, it remains of interest whether bacteria follow a similar degradation scheme to the NRD in eukaryotes. Escherichia coli has been the subject of the majority of prokaryote study. Numerous differences were observed between the degradation of eukaryotic and prokaryotic rRNA, leading researchers to conclude that the two types of rRNA follow distinct degradation pathways.
- Certain rRNA mutations that triggered rRNA degradation in eukaryotes were incapable of doing so in prokaryotes.
- In prokaryotes, point mutations in a 23S rRNA would result in the degradation of both the 23S and 16S rRNAs, whereas in eukaryotes, mutations in one subunit would result in the degradation of only that subunit.
- The removal of a complete helix structure (H69) from the 23S rRNA did not result in its deterioration, as discovered by researchers. They concluded that H69 was essential for endonucleases to recognise and degrade mutated rRNA.
Functions of rRNA
- Protein synthesis is rRNA’s primary function. The ribosome’s A, P, and E sites are generated by the peculiar three-dimensional structure of rRNA, which consists of internal helices and loops.
- By binding to messenger RNA and transfer RNA, these molecules ensure that the codon sequence of the mRNA is correctly translated into the amino acid sequence of the protein.
- The A site anchors an entering tRNA that has been charged with an amino acid, whereas the P site is responsible for binding a nascent polypeptide. Following the formation of a peptide bond, the tRNA momentarily adheres to the E site before leaving the ribosome.
- In addition, some ribosomal proteins can bind to rRNA at specific residues, which have been identified by in-depth analyses of both the RNA and protein.
- Recent research has revealed that antibiotics such as streptomycin and tetracycline have binding sites on bacterial rRNA. For instance, Euglena and Escherichia coli’s tolerance to streptomycin results from a mutation in the 16S rRNA sequence.
- It appears that the 30S rRNA is the cause of tetracycline resistance. Similar results were discovered for Streptomyces’ resistance to spectinomycin.
- Preribosomal RNA, one of rRNA’s ancestors, has been connected to the creation of microRNA, which mediates mechanical stress-related inflammation and heart disease. This discovery adds a new dimension to rRNA’s function.
Significance of rRNA
The features of ribosomal RNA are essential to evolution, and consequently to taxonomy and medicine.
- rRNA is one of the few gene products that are present in every cell. Therefore, genes encoding rRNA (rDNA) are sequenced to determine the taxonomic category of an organism, calculate related groups, and estimate rates of species divergence. Consequently, tens of thousands of rRNA sequences are known and maintained in specialist databases like RDP-II[65] and SILVA.
- Certain disease-causing bacteria, such as Mycobacterium TB (the bacterium that causes tuberculosis), develop high treatment resistance due to modifications to rRNA. Due to similar concerns, this has become a prevalent issue in veterinary medicine, where the primary treatment for bacterial infections in dogs is the administration of medications that target the peptidyl-transferase centre (PTC) of the bacterial ribosome. Mutations in 23S rRNA have developed perfect resistance to these medications, which work in an unknown manner to circumvent the PTC.
- Numerous clinically significant antibiotics target rRNA, including chloramphenicol, erythromycin, kasugamycin, micrococcin, paromomycin, ricin, alpha-sarcin, spectinomycin, streptomycin, and thiostrepton.
- It has been demonstrated that rRNA is the source of species-specific microRNAs, such as miR-663 in humans and miR-712 in mice. These specific miRNAs are derived from the internal transcribed spacers of rRNA.
References
- Malhotra, A. (2001). rRNA Structure. Encyclopedia of Life Sciences. doi:10.1038/npg.els.0000537
- Brimacombe R, Stiege W. Structure and function of ribosomal RNA. Biochem J. 1985 Jul 1;229(1):1-17. doi: 10.1042/bj2290001. PMID: 3899100; PMCID: PMC1145144.
- Noller, H. F., Green, R., Heilek, G., Hoffarth, V., Hüttenhofer, A., Joseph, S., … Weiser, B. (1995). Structure and function of ribosomal RNA. Biochemistry and Cell Biology, 73(11-12), 997–1009. doi:10.1139/o95-107
- https://serc.carleton.edu/microbelife/research_methods/genomics/ribosome.html
- https://www.rcsb.org/structure/1pbr
- https://www.vedantu.com/biology/ribosomal-rna
- https://microbe.net/simple-guides/fact-sheet-ribosomal-rna-rrna-the-details/
- http://www.actabp.pl/pdf/1_2001/191.pdf
- https://www.news-medical.net/life-sciences/-Types-of-RNA-mRNA-rRNA-and-tRNA.aspx
- https://biologydictionary.net/ribosomal-rna/
- https://en.wikipedia.org/wiki/Ribosomal_RNA
