Table of Contents
What is a vector in biology?
- A vector, in the context of genetic engineering, is a vital tool used to transfer genetic material from one cell to another. It acts as a carrier, introducing a specific DNA sequence or other genetic material into a new cell. Vectors play a crucial role in various molecular cloning procedures, facilitating the insertion of desired DNA fragments into host cells.
- Vectors are typically DNA sequences that consist of different components serving different functions. They contain an insert, also known as a transgene, which carries the recombinant DNA to be transferred. Additionally, vectors possess a larger sequence called the backbone, which provides structural support to the vector.
- Classification of vectors depends on specific characteristics, and the choice of vector depends on the intended purpose of the genetic engineering process. Vectors are fundamental in genetic engineering as they form the basis for transferring DNA fragments between cells. They possess unique features that allow the gene sequences to survive within the host cell.
- The process of gene transfer can vary depending on the type of vector used. Some vectors integrate into the host DNA after entering the cell, while others solely pass the genetic material to the host cell without integrating themselves. Additionally, besides DNA sequences, viruses and other particles can also serve as vectors in processes such as transduction.
- Vectors can be reused in multiple processes as they can be recovered at the end of each procedure. Cloning vectors, specifically designed for cloning purposes, are a specific category of vectors. These vectors contain sequences that enable them to initiate replication within host cells and propagate themselves effectively.
- In summary, vectors are essential tools in genetic engineering that enable the transfer of genetic material between cells. They play a vital role in molecular cloning procedures, facilitating the insertion of desired DNA fragments. Understanding the characteristics and types of vectors is crucial for effective genetic engineering processes.
Vector Definition
A vector is a DNA molecule or genetic material used to transfer specific genetic information between cells in molecular biology and genetic engineering.
Characteristics Features of vectors
Vectors used in molecular biology and genetic engineering possess certain characteristic features that make them suitable for specific applications. Here are some key characteristics of commonly used vectors:
- Plasmids: Plasmid vectors are circular, double-stranded DNA molecules that can replicate independently within host cells. They often contain an origin of replication, which allows for the semi-independent replication of the plasmid within the host. Plasmids can be found in bacteria and some eukaryotes like yeast. They are widely used for cloning purposes and can be easily manipulated in the laboratory. Plasmids used as cloning vectors may have a “multiple cloning site” where DNA fragments can be inserted, and they can also serve as transcription vectors or expression vectors for protein production.
- Viral Vectors: Viral vectors are genetically engineered viruses that have been modified to carry and deliver foreign DNA or RNA into host cells. They are rendered noninfectious but retain viral promoters and transgene sequences, allowing for the expression of the transgene in the host cell. Viral vectors can efficiently transfer genetic material into a wide range of cell types and have the potential for long-term expression. However, they require helper viruses or packaging lines for large-scale transfection. Viral vectors, such as retroviruses, leave specific genetic markers in the host genome after integrating the transgene.
- Artificial Chromosomes: Artificial chromosomes are engineered chromosome-like structures used to carry large DNA fragments. They can be categorized into yeast artificial chromosomes (YACs), bacterial artificial chromosomes (BACs), or human artificial chromosomes (HACs). YACs and BACs have the ability to accommodate DNA fragments up to hundreds of thousands of nucleotides long. They contain essential components, including an origin of replication, a centromere, and telomeric end sequences, to ensure stable replication and segregation during cell division.
These vectors offer distinct advantages and are chosen based on the specific requirements of the experiment or application. Plasmid vectors are versatile and widely used for cloning and protein expression studies. Viral vectors, although more complex, are efficient in delivering genetic material into cells and enable long-term gene expression. Artificial chromosomes are valuable for carrying large DNA fragments, making them suitable for genome mapping and studying complex genetic elements.
The selection of a vector depends on factors such as the size of the DNA insert, the target host organism, the desired level and duration of gene expression, and the downstream applications. By utilizing the unique characteristics of these vectors, researchers can manipulate and study genetic material with precision, advancing our understanding of biology and enabling various biotechnological applications.
Types of vectors
Vectors are classified into these following types;
- Cloning Vectors: Cloning vectors are specialized vectors used in molecular cloning procedures. These vectors have specific features that allow for the replication and propagation of DNA fragments within host cells. They typically contain essential elements such as an origin of replication, selectable markers, and cloning sites to facilitate the cloning and manipulation of DNA fragments.
- Viral Vectors: Viral vectors are vectors derived from viruses and are widely used in gene therapy and gene delivery applications. These vectors are engineered to carry and deliver therapeutic genes or desired genetic material into target cells. Viral vectors utilize the natural ability of viruses to infect cells, making them efficient delivery vehicles for introducing genetic material into host cells.
- Expression Vector: Expression vectors are designed to facilitate the expression of genes in host cells. These vectors typically contain regulatory elements such as promoters, enhancers, and terminators that control gene expression. Expression vectors are widely used in molecular biology research to produce proteins of interest or study gene function by inducing the expression of specific genes in host cells.
- Shuttle Vector: Shuttle vectors are versatile vectors that can replicate in multiple host organisms. These vectors possess sequences allowing them to be maintained in different host species, such as bacteria and eukaryotic cells. Shuttle vectors enable the cloning and manipulation of DNA fragments in one host system and subsequent transfer into another host system for further analysis or expression.
- Secretion Vector: Secretion vectors are designed to facilitate the secretion of proteins or peptides of interest into the extracellular space. These vectors contain signal sequences that direct the protein to be synthesized and secreted from the host cell. Secretion vectors are commonly used in biotechnology and protein production applications, as they allow for easy recovery and purification of the secreted proteins.
1. Cloning vectors
- A cloning vector, an essential tool in molecular biology, is a small piece of DNA that allows the insertion of foreign DNA fragments for cloning purposes. Cloning vectors can be derived from various sources, such as viruses, higher organisms, or bacterial plasmids. They possess specific features that facilitate the insertion and removal of DNA fragments, typically through the presence of restriction sites.
- The most commonly used cloning vectors are genetically engineered plasmids, and they are often employed in Escherichia coli (E. coli) bacteria. These vectors, including plasmids, bacteriophages like phage λ, cosmids, and bacterial artificial chromosomes (BACs), contain elements necessary for propagation and maintenance in E. coli, such as an origin of replication (ori). Some cloning vectors, known as shuttle vectors, can be maintained in multiple organisms, including E. coli.
- Key features of cloning vectors include a suitable cloning site and selectable marker. The cloning site, often referred to as a multiple cloning site (MCS) or polylinker, contains multiple unique restriction sites. These sites are cleaved by restriction enzymes, allowing the insertion of a PCR-amplified target gene digested with the same enzymes. Cloning vectors may also utilize alternative methods, such as topoisomerase or DNA recombination, to facilitate cloning without the need for restriction digest and ligation.
- Selectable markers carried by the cloning vectors enable the selection of positively transformed cells. Common markers include antibiotic resistance genes, like the beta-lactamase gene conferring resistance to penicillin. Some vectors have dual selectable markers, while others utilize auxotrophic selection markers for specific organisms. Additionally, certain cloning vectors incorporate reporter genes, such as lacZα or marker genes, to aid in the identification of successful clones.
- Expression vectors, a subset of cloning vectors, contain elements for gene expression, including promoters and ribosomal binding sites (RBS). These vectors allow the cloned target gene to be expressed under the control of a specific promoter, ensuring tight regulation and inducible expression. Prominent promoters used in expression vectors include T7 and lac promoters.
- While some cloning vectors lack expression elements, they are still valuable for cloning genes toxic to E. coli cells or for creating genomic or cDNA libraries. Transcription vectors are specialized cloning vectors designed solely for transcription without producing heterologous proteins, primarily used for in vitro mRNA production.
- Cloning vectors are indispensable tools in molecular biology research, enabling the cloning and manipulation of DNA fragments, the creation of genomic libraries, and the production of recombinant proteins. Their diverse features and versatility make them vital for a wide range of applications in genetic engineering and biotechnology.
Types of cloning vectors
a. Plasmid vector
- Plasmid vectors play a pivotal role in recombinant DNA technology as versatile tools for cloning and genetic engineering. These small circular DNA molecules are capable of autonomous replication within host cells, making them highly valuable in molecular biology research. Plasmids are widely utilized as vectors across various organisms, with a particular emphasis on bacteria and yeasts.
- One of the primary advantages of plasmid vectors is their compact size. This small size allows for efficient separation of the recombinant DNA from the host’s genomic DNA. Plasmids typically range in size from a few thousand base pairs to over 100 kilobases, although the maximum size of the insert DNA they can carry is influenced by their own size. For optimal cloning efficiency and plasmid stability, insert DNA should generally be less than 20 kb.
- Plasmids owe their replicative capability to specific genes and sequences that enable them to initiate replication independently of the host’s replication cycle. Bacterial plasmids contain origin of replication (ori) sequences that govern plasmid replication and influence the coexistence of multiple plasmids within the same host cell.
- Plasmids exhibit various selective markers, with antibiotic resistance and β-galactosidase enzyme production being among the most common. Selective markers allow for the identification and selection of cells that have successfully taken up the plasmid vector.
- Several well-known plasmid vectors are widely employed in research. The pBR322, pUC, and pBluescript vectors, utilizing Escherichia coli (E. coli) as the host, are examples of extensively used plasmids. These vectors offer specific advantages and features tailored to different applications.
- In summary, plasmid vectors serve as indispensable tools in recombinant DNA technology. Their compact size, autonomous replication ability, and diverse selective markers make them versatile and widely applicable in various domains of life. Plasmid vectors, especially those designed for use in bacteria like E. coli, have revolutionized genetic engineering and continue to drive advancements in biotechnology and molecular biology research.
Examples of Plasmid Vectors:
- pUC19: A widely used cloning vector with a high copy number and multiple cloning sites.
- pBR322: One of the first plasmid vectors used for cloning, containing antibiotic resistance genes.
- pGEM-T: A vector for cloning PCR products using T/A cloning strategy.
b. Bacteriophage vector
- Bacteriophage vectors, specifically the λ phage and M13 phage, are powerful tools in molecular cloning. These viruses exclusively infect bacteria and efficiently transform them while accommodating large DNA inserts. Bacteriophages offer distinct advantages, including high transformation efficiencies that increase the likelihood of recovering clones containing recombinant DNA segments.
- One crucial feature of bacteriophages is their packaging system, which enables the incorporation of substantial eukaryotic genes along with their regulatory elements. This characteristic facilitates the isolation of larger quantities of DNA, which can be subsequently used for insert analysis. Among the available bacteriophages, phage λ is the most commonly used and convenient cloning vector. It selectively packages a chromosome approximately 50 kb in length, and the phage size can be adjusted by removing the central part of its genome, as it is unnecessary for replication or packaging donor DNA.
- Using a bacteriophage vector capable of incorporating larger DNA segments reduces the number of clones required to construct a DNA library representing the entire genome of an organism. Furthermore, phage vectors are efficient cloning vectors, as the recombinant molecules formed during the cloning process are packaged into infective particles that can be stored and handled with ease.
- Several bacteriophages have been employed as vectors, including M13 phages, λ phages, and P1 phages. Each of these vectors offers specific advantages and may be suitable for different cloning applications.
- In summary, bacteriophage vectors are valuable tools in molecular cloning, particularly for accommodating large DNA inserts. Their efficient transformation capabilities, ability to package recombinant molecules, and capacity to incorporate substantial genetic material make them indispensable in biotechnology and genetic engineering research.
c. Yeast artificial chromosome
- Yeast artificial chromosomes (YACs) serve as essential vectors for cloning large DNA fragments, surpassing 1 megabase (1Mb = 1000kb) in size. These vectors play a crucial role in genome mapping projects like the Human Genome Project, where cloning larger DNA fragments is necessary. YACs contain specific features, including a telomeric sequence and an autonomously replicating sequence, required for replicating linear chromosomes in yeast cells. Additionally, these vectors incorporate suitable restriction sites to clone foreign DNA and genes used as selectable markers.
- Yeast artificial chromosomes are engineered DNA molecules designed to clone DNA inserts within yeast cells, particularly Saccharomyces cerevisiae. They have been developed to enhance the efficiency of cloning by accommodating large DNA sequences. YACs are capable of cloning up to 500 kb of DNA, surpassing the capacity of most traditional cloning vectors. While commonly used as cloning vectors, they also find utility in DNA sequencing and analysis, as well as in cloning complete sequences of larger genomes that exceed the limits of traditional techniques.
- One distinct advantage of YACs is their ability to clone unstable sequences when using prokaryotic systems, as yeast cells are eukaryotic. YACs consist of functional units from various organisms and, once the insert DNA is cloned, they function as replicating yeast chromosomes. However, there are limitations associated with using YACs as vectors, such as a high degree of chimerism and insert rearrangement. Furthermore, due to their eukaryotic nature, yeast cells are more challenging to handle and have lower efficiencies compared to bacterial artificial chromosomes.
- Over the years, different types of yeast artificial chromosomes have been created to serve various purposes. One commonly used example is pYAC4, extensively employed as a cloning vector.
- In summary, yeast artificial chromosomes (YACs) are powerful vectors for cloning large DNA fragments, exceeding 1 megabase in size. They offer advantages in genome mapping and cloning projects, providing the capacity to clone extensive DNA sequences. While presenting some limitations, YACs play a crucial role in genetic research, allowing the cloning of unstable sequences and the analysis of larger genomes. pYAC4 stands as a prominent example of a widely used yeast artificial chromosome vector.
Examples of Yeast Artificial Chromosomes
- pYAC4: A YAC vector used for cloning large DNA fragments in yeast.
- pCYPAC2: A YAC vector used for constructing genomic libraries and functional analysis of large DNA segments.
d. Cosmid
- Cosmids are hybrid vectors that combine elements of plasmids and bacteriophage λ vectors. They incorporate a segment of bacteriophage λ DNA containing the cohesive end site (cos), which includes the necessary elements for packaging DNA into λ particles. These vectors are primarily used for cloning large DNA fragments ranging from 28 to 45 kilobases (Kb) in size. Cosmid vectors possess the ability to incorporate up to 42 kb of DNA and are prepared by inserting the cos region of the phage vector into the plasmid vectors.
- Cosmid vectors are relatively large, ranging in size from 400 base pairs to 30 kb. They can accommodate DNA sequences of varying sizes, typically ranging from 28 to 46 kb. These vectors were developed to enable the cloning of larger DNA molecules that cannot be efficiently carried by plasmids alone.
- The hybrid nature of cosmid vectors allows them to replicate within the host cell similar to plasmids or remain packaged like a phage. However, cosmid vectors do not exhibit many phage characteristics except for the presence of signal sequences that promote phage-head packaging. The unique structure of cosmid vectors facilitates the incorporation of phage heads into all donor DNA during transfer.
- Over the years, the use and production of cosmid vectors have increased due to their highly efficient and selective recovery of larger hybrid constructs. These vectors offer a robust packaging system for larger DNA molecules. An example of a commonly used cosmid vector is cosmid pHC79, which is a derivative of the vector pBR322 and contains the cos region.
- In summary, cosmids are hybrid vectors that combine plasmid and phage λ elements. They allow for the cloning of large DNA fragments and are prepared by incorporating the cos region of the phage vector into plasmid vectors. Cosmid vectors have a diverse size range and can carry substantial DNA sequences. Their hybrid structure provides efficient and selective recovery of larger DNA constructs. Cosmid pHC79 serves as a practical example of a cosmid vector widely used in research and applications.
Examples of Cosmid Vectors
- SuperCos1: A cosmid vector derived from the lambda phage, used for the cloning and analysis of large DNA fragments up to 45 kilobases (kb).
- pWE15: A cosmid vector with a large insert capacity of up to 50 kb, commonly used for genomic library construction and functional studies.
- pEPI-1: A cosmid vector used for the preparation of large DNA fragments and for cloning unstable DNA sequences.
e. Human artificial chromosome
- Human artificial chromosomes (HACs) are extrachromosomal DNA fragments that function as independent chromosomes within human cells. These artificial chromosomes have emerged as potentially valuable tools in gene transfer vectors for delivering genes into human cells, as well as for expression studies and investigating human chromosome function. One of the significant advantages of HACs is their ability to carry very large DNA fragments without a practical upper size limit. This overcomes the limited cloning capacity found in other vectors and eliminates the potential risks of insertional mutagenesis that can arise from integration into host chromosomes, as seen with viral vectors.
- The use of human artificial chromosomes has gained momentum alongside advancements in genetic engineering, offering solutions to challenges commonly associated with traditional vector systems. HACs can exist as single-copy episomes within cells, meaning they are not integrated into the host chromosomes. This characteristic allows for long-term stable maintenance of the artificial chromosomes. Additionally, HACs provide the flexibility to incorporate DNA inserts of any size, including entire genomic units, enabling the mimicry of natural gene expression.
- Although HACs offer numerous advantages, their utilization has been largely limited to studies focused on understanding the structure and function of human kinetochores. This is due, in part, to technical difficulties encountered during gene loading and the relatively ill-defined structures of the vectors themselves.
- In summary, human artificial chromosomes (HACs) are extrachromosomal DNA fragments that function as separate chromosomes within human cells. They offer significant advantages as gene transfer vectors, overcoming limitations in cloning capacity and avoiding potential risks associated with viral vector integration. HACs can carry large DNA fragments without a practical size limit and can be stably maintained as episomes. However, their applications have primarily been focused on studying human kinetochores, and challenges related to gene loading and vector structures remain to be addressed.
f. Bacterial artificial chromosome
- Bacterial artificial chromosomes (BACs) are engineered DNA molecules utilized for cloning DNA segments within bacterial cells, typically Escherichia coli (E. coli). These vectors incorporate a replication origin derived from the F-factor in bacteria, enabling the propagation of large DNA fragments in a supercoiled circular form.
- One of the primary advantages of BACs is their capacity to carry significantly larger insert DNA sizes compared to plasmid or phage vectors. With insert sizes of up to 350 kilobases (kb), BACs provide ample space for cloning large DNA segments. This feature reduces the number of clones and cycles required to achieve desired outcomes, enhancing efficiency in genetic engineering experiments.
- Bacterial artificial chromosomes are particularly favored over other artificial chromosomes such as yeast artificial chromosomes and mammalian artificial chromosomes due to the inclusion of the F-factor derived from bacteria. This F-factor helps minimize insert chimerism and instability that may arise during the cloning process, ensuring greater stability and fidelity.
- BAC libraries have proven invaluable in generating large genomic DNA inserts, facilitating processes like positional cloning, physical mapping, and genome sequencing. The stability and ease of use associated with the BAC cloning system have contributed to its increasing adoption in genetic engineering.
- However, it is worth noting that BACs have been associated with the potential for random insertion of DNA fragments into the host genome, which can lead to unpredictable gene expression patterns. Careful consideration and analysis are necessary to minimize the impact of this phenomenon in experimental designs involving BAC vectors.
- In summary, bacterial artificial chromosomes (BACs) are engineered DNA molecules employed for cloning DNA segments in bacterial cells. BACs offer a larger insert capacity compared to other vectors, reducing the number of clones required for successful outcomes. The inclusion of the F-factor derived from bacteria enhances stability, making BACs an attractive choice in genetic engineering applications. Nonetheless, caution is needed to mitigate potential issues related to random DNA fragment insertion into the host genome, which can result in unanticipated gene expression patterns.
Examples of Bacterial Artificial Chromosomes (BACs)
- pBeloBAC11: A widely used BAC vector for cloning large DNA fragments up to 300,000 base pairs.
- pBACe3.6: A BAC vector used for constructing genomic libraries and large-scale DNA sequencing.
g. Animal and plant viral vectors
- Animal and plant viral vectors have been extensively utilized in genetic engineering to introduce foreign genes into animal and plant cells. These vectors take advantage of the natural properties of viruses, such as their ability to adsorb to cells, introduce their DNA, and replicate within the host cells. This characteristic makes viruses ideal vehicles for transferring foreign DNA into eukaryotic cells in culture.
- The pioneering work in mammalian cell cloning experiments involved the use of a vector based on Simian virus 40 (SV40). This vector demonstrated the potential of viral vectors in introducing foreign genes into mammalian cells. Since then, various viral vectors based on different types of viruses, such as Adenoviruses and Papillomaviruses, have been employed to clone genes in mammals. Currently, retroviral vectors have gained popularity as effective tools for gene cloning in mammalian cells.
- In the realm of plant genetic engineering, viral vectors have also been explored, although with limited success compared to their animal counterparts. Viruses like Cauliflower mosaic virus, Tobacco mosaic virus, and Gemini viruses have been utilized in attempts to clone genes in plants. While progress has been made, there are still challenges to overcome in achieving efficient gene transfer and expression in plant cells using viral vectors.
- The versatility and efficiency of animal and plant viral vectors make them valuable tools in genetic engineering. Their ability to infect cells, introduce foreign DNA, and exploit the host’s replication machinery allows for the successful transfer and expression of desired genes. Ongoing research aims to refine and optimize these viral vectors to enhance their efficiency, safety, and specificity in delivering genes into animal and plant cells.
- In summary, animal and plant viral vectors have revolutionized genetic engineering by enabling the transfer of foreign genes into animal and plant cells. These vectors leverage the natural properties of viruses to efficiently deliver DNA into host cells. While viral vectors have been extensively utilized in mammalian cell cloning experiments, their application in plant genetic engineering is still evolving. Continued research and advancements in viral vector technology hold promise for further enhancing their utility in diverse genetic engineering applications.
2. Viral vectors
- Viral vectors have emerged as highly effective tools for gene transfer, allowing the modification of host cells and tissues to express a wide range of genes. The concept of using viruses as vectors stems from their inherent ability to efficiently deliver their genetic material into host cells.
- Viral transduction involves the modification of viral genomes by replacing non-essential viral genes with foreign DNA sequences of therapeutic interest. This process results in the generation of recombinant viral vectors capable of delivering the desired genes to target cells.
- Researchers have extensively studied various groups of viruses for their potential use as viral vectors, offering transient or permanent transgene expression. Commonly investigated virus groups include adenoviruses, retroviruses, poxviruses, and adeno-associated viruses. The selection of a specific virus as a vector depends on several factors, such as the efficiency of transgenic expression, ease of production, safety considerations, and vector stability.
- Viral vectors provide the advantage of location specificity through unique injection techniques, allowing for targeted delivery of genes to specific tissues or cells within defined timeframes. This specificity enhances the precision and effectiveness of gene transfer.
- Numerous clinical trials have been conducted using diverse viral vectors tailored for specific applications. For instance, adenoviruses have been employed for transferring tumor suppressor genes in cancer treatment, while retroviruses are being explored for their potential in tissue repair and engineering.
- The continuous advancement of viral vector technology holds great promise for gene therapy and other genetic engineering applications. Ongoing research aims to refine and improve vector efficiency, safety profiles, and scalability of production.
- In summary, viral vectors have proven to be powerful tools in gene transfer, allowing for the manipulation of host cells to express desired genes. Different groups of viruses have been investigated for their potential as viral vectors, each offering unique characteristics and advantages. The selection of a specific viral vector depends on factors such as transgenic expression efficiency, production ease, safety considerations, and stability. As research progresses, viral vectors hold significant potential for advancing gene therapy and other therapeutic interventions.
Examples of Viral Vectors
- Lentiviral Vectors: Derived from the human immunodeficiency virus (HIV), used for stable gene transfer in both dividing and non-dividing cells.
- Adenoviral Vectors: Derived from adenoviruses, commonly used for efficient gene delivery to various cell types.
- Retroviral Vectors: Derived from retroviruses, used for integrating genes into the host genome and long-term gene expression.
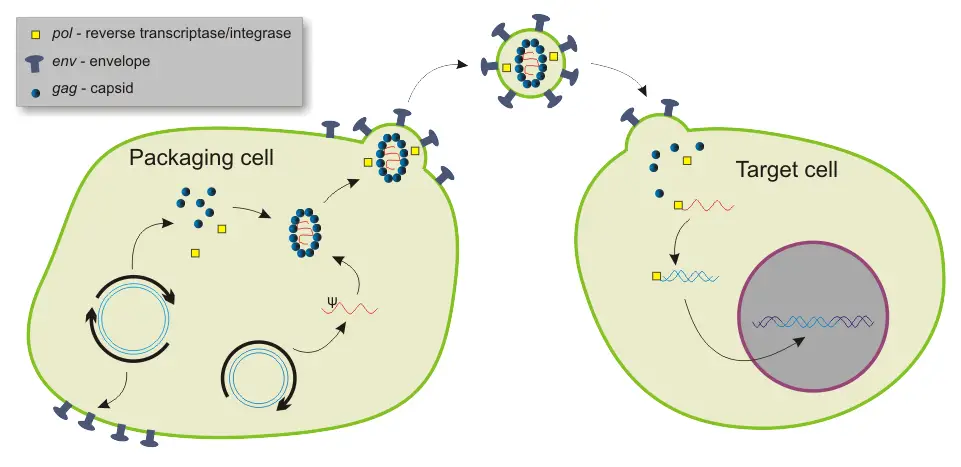
3. Shuttle vector
- Shuttle vectors are versatile tools used in molecular biology research to transfer DNA between different host organisms. These vectors carry origins of replication from two distinct hosts, allowing them to “shuttle” or move between these hosts.
- Shuttle vectors are typically DNA plasmids that have the ability to replicate in both bacterial and mammalian cells. They serve as hybrid vectors, incorporating DNA sequences from bacterial plasmids and mammalian viruses. These vectors play a crucial role in cloning processes and gene transfer experiments.
- The design of shuttle vectors includes three essential functional DNA sequences: a viral replication origin, a bacterial replication origin, and a drug resistance gene. The presence of these elements enables efficient replication and maintenance of the vectors in bacterial cells.
- There are three main types of shuttle vectors, each utilizing a different replication system. Transiently replicating shuttle vectors require recognition by large T antigen in order to replicate within human cells. These vectors are commonly used for short-term experiments.
- Episomal shuttle vectors, on the other hand, are designed to establish cell lines that can permanently replicate the plasmid DNA containing the desired DNA insert. These vectors provide a stable long-term expression of the inserted genes.
- Integrated shuttle vectors function differently as they undergo replication only after fusion with specific cell types, facilitating targeted gene expression in those cells.
- Shuttle vectors have revolutionized genetic engineering research by enabling the transfer of DNA between different host organisms. They have facilitated the study of gene function, gene therapy development, and the production of recombinant proteins. The ability of these vectors to shuttle between hosts has expanded the range of experiments and applications in molecular biology.
- In summary, shuttle vectors are valuable tools in molecular biology research, allowing DNA transfer between different host organisms. They consist of plasmids with origins of replication from both bacterial and mammalian cells. With their distinct replication systems, shuttle vectors offer flexibility in experimental design and facilitate the study of gene function and therapeutic applications.
Examples of Shuttle Vectors
- pUC18: A widely used shuttle vector that can replicate in both Escherichia coli and other Gram-negative bacteria.
- pRS416: A shuttle vector commonly used in Saccharomyces cerevisiae for cloning and expression of genes.
- pCMV6-AC: A shuttle vector designed for mammalian cell expression, containing a cytomegalovirus (CMV) promoter for high-level gene expression.
4. Expression vector
- An expression vector, also known as an expression construct, is a powerful tool used in biotechnology to facilitate gene expression in cells. Typically in the form of a plasmid or virus, an expression vector is designed to introduce a specific gene into a target cell and enable the production of the protein encoded by that gene. These vectors play a fundamental role in protein production and are widely used in various fields of research and industrial applications.
- Expression vectors are meticulously engineered to include specific regulatory sequences that act as enhancers and promoters, essential for efficient transcription of the gene carried on the vector. The ultimate goal of a well-designed expression vector is to achieve high levels of protein production. This is often accomplished by ensuring the generation of a significant amount of stable messenger RNA (mRNA), which can then be translated into protein. The expression of a protein can be tightly controlled, allowing it to be produced only when needed, through the use of an inducer. In certain systems, however, the protein may be expressed constitutively, meaning it is constantly produced. The bacterium Escherichia coli (E. coli) is a common host organism used for protein production, although other cell types can also be employed. An exemplary application of expression vectors is the production of insulin, a crucial protein used in the medical treatment of diabetes.
- An expression vector possesses several key elements commonly found in other types of vectors. These elements include an origin of replication, a selectable marker, and a suitable site for the insertion of the gene, often referred to as the multiple cloning site. The cloned gene can be transferred from a specialized cloning vector to the expression vector, although it is also possible to directly clone into an expression vector. The cloning process typically takes place in E. coli. In cases where protein production is desired in organisms other than E. coli, shuttle vectors come into play. These vectors contain additional elements that allow them to be maintained in the desired organism, alongside a suitable origin of replication for propagation in E. coli.
- Crucial to the success of gene expression, an expression vector must incorporate elements necessary for gene expression, including a promoter, a translation initiation sequence (such as a ribosomal binding site and start codon), a termination codon, and a transcription termination sequence. Differences exist in the machinery responsible for protein synthesis between prokaryotes and eukaryotes, necessitating the use of appropriate expression elements specific to the chosen host organism. For example, prokaryotic expression vectors include a Shine-Dalgarno sequence at the translation initiation site to facilitate ribosome binding, while eukaryotic expression vectors often contain the Kozak consensus sequence.
- The promoter is a crucial element as it initiates transcription and serves as a point of control for the expression of the cloned gene. Promoters used in expression vectors are typically inducible, meaning that protein synthesis is only triggered when an inducer, such as IPTG, is introduced. However, some expression vectors may utilize constitutive gene expression, resulting in continuous protein production. Even with tightly controlled promoters, low levels of constitutive protein synthesis may still occur.
- To facilitate protein purification, expression vectors may incorporate protein tags. These tags, such as histidine (His) tags or fusion partners like glutathione S-transferase or maltose-binding protein, simplify the separation of the protein of interest from the majority of host cell proteins during purification. Additionally, certain fusion partners can enhance the solubility of expressed proteins. Other fusion proteins, such as green fluorescent protein, can serve as reporter genes to identify successful cloning or be utilized for studying protein expression through cellular imaging.
- Expression vectors are transformed or transfected into host cells to initiate protein synthesis. Some vectors may contain elements for transformation or DNA insertion into the host chromosome, such as the vir genes used for plant transformation or integrase sites for chromosomal integration. Furthermore, certain vectors may incorporate targeting sequences that direct the expressed protein to specific locations within the cell, such as the periplasmic space of bacteria.
- In summary, expression vectors are essential tools in biotechnology that enable gene expression and protein production in cells. They are carefully designed with regulatory sequences to ensure efficient transcription and translation of the cloned genes. By utilizing expression vectors, researchers can manipulate the cellular machinery to produce specific proteins of interest, enabling a wide range of applications in research, medicine, and various industries.
Examples of Expression Vectors
- pET vectors: A series of vectors commonly used for protein expression in Escherichia coli (E. coli) with inducible promoters and fusion tags for protein purification.
- pDEST vectors: Gateway cloning compatible vectors used for expression in multiple systems, such as E. coli, yeast, and mammalian cells.
5. Secretion vector
- Secretion vectors are specialized expression vectors that allow for the production of proteins at locations other than the cytoplasm, enabling the secretion of the protein product from the host cell. These vectors are designed to transport the protein of interest out of the cell by fusing the inserted DNA with a nucleotide sequence encoding a signal peptide or other secretion-related elements.
- The utilization of secretion vectors offers several advantages in protein production. Firstly, it enables higher protein yields compared to traditional cytoplasmic expression. By directing the protein to be secreted, larger quantities can be obtained, which is particularly beneficial for industrial-scale production.
- Moreover, secretion vectors simplify the purification process. Since the protein of interest is secreted into the extracellular space or culture medium, it can be isolated and purified from the surrounding environment rather than from the complex cellular mixture. This streamlined purification process saves time and resources.
- In addition, secretion vectors contribute to improved protein stability. The extracellular environment often provides more favorable conditions for protein folding and stability, reducing the risk of aggregation or degradation. This can result in higher-quality proteins with enhanced functionality.
- Secretion vectors can be designed for use in various host systems, including prokaryotes and eukaryotes, such as mammalian cells. These vectors can be engineered to accommodate the specific requirements of different expression systems, allowing for versatile applications.
- One common challenge when expressing eukaryotic proteins in prokaryotic hosts is the formation of inclusion bodies, which are aggregates of improperly folded proteins. Secretion vectors help overcome this issue by directing the protein to be secreted, minimizing the formation of inclusion bodies and increasing the likelihood of obtaining soluble, functional proteins.
- As a result, secretion vectors have become essential tools in protein production and the expression of eukaryotic DNA fragments. They have largely replaced traditional cloning vectors in processes that focus on generating recombinant proteins for various applications, including biopharmaceuticals, industrial enzymes, and research tools.
- In summary, secretion vectors provide an efficient and optimized approach for protein production by enabling the secretion of proteins from host cells. Their advantages include higher yields, simplified purification processes, improved protein stability, and the ability to overcome challenges associated with expressing eukaryotic proteins in prokaryotic hosts. These vectors have revolutionized the field of protein expression and have become indispensable in various biotechnological and biomedical applications.
Examples of Secretion Vectors
- pPICZ: A secretion vector used in the yeast Pichia pastoris system for the production and secretion of recombinant proteins.
- pSecTag: A mammalian secretion vector that enables the secretion of recombinant proteins into the culture medium.
- pBAD/His: A secretion vector that allows inducible expression and secretion of recombinant proteins in Escherichia coli (E. coli) using the arabinose promoter.
Examples of Vectors
pUC19
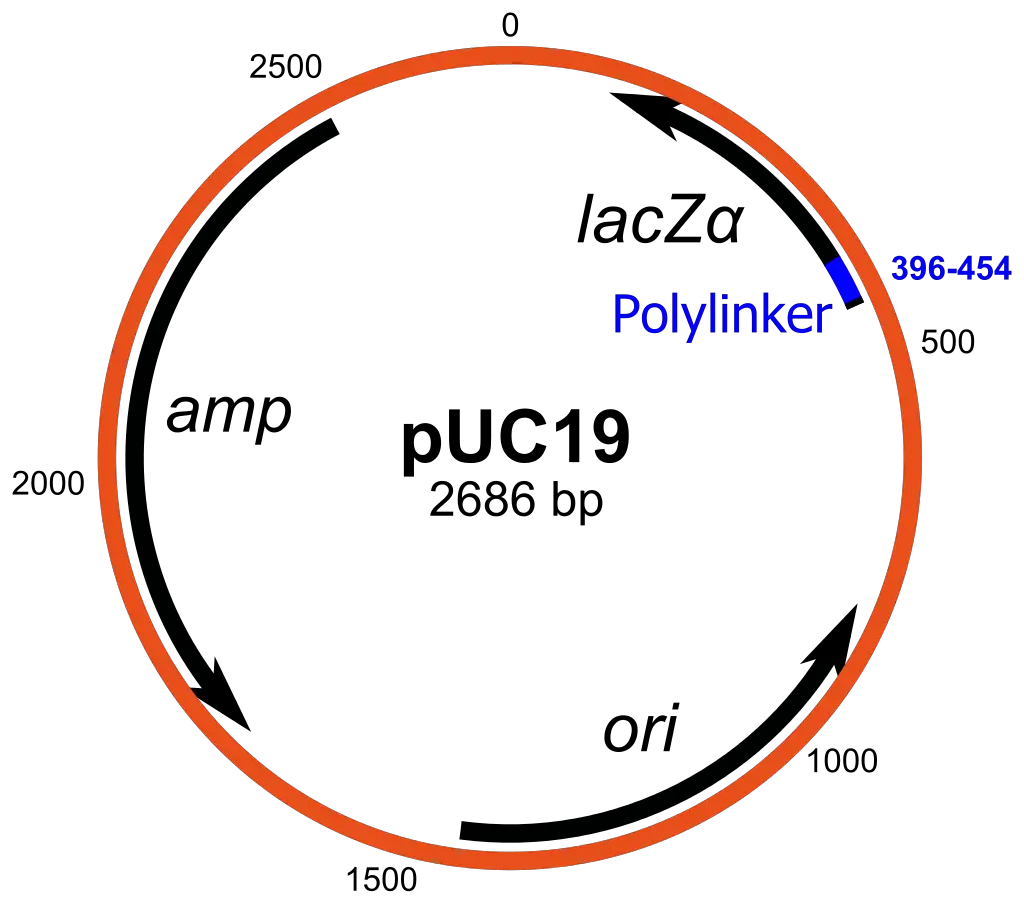
- pUC19 is a widely used plasmid cloning vector in molecular biology research. Created by Joachim Messing and his team, the name “pUC” represents “plasmid” and the abbreviation for the University of California, where the vector was developed. With a circular double-stranded DNA structure, pUC19 consists of 2686 base pairs, making it compact and efficient for cloning purposes. This vector is highly popular due to its ability to distinguish between recombinant and non-recombinant cells based on colony color variations on growth media.
- One of the key features of pUC19 is its inclusion of the N-terminal fragment of the β-galactosidase (lacZ) gene from E. coli. This gene serves as a marker for screening recombinant cells. The multiple cloning site (MCS) region within pUC19 is strategically positioned between codons 6-7 of the lacZ gene, allowing for the insertion of foreign DNA at numerous restriction endonuclease sites. Additionally, pUC19 carries an ampicillin resistance gene (ampR) that encodes the β-lactamase enzyme, which confers resistance to ampicillin by degrading the antibiotic and protecting the host cell.
- The origin of replication (ori) in pUC19 is derived from the plasmid pMB1. Despite its small size, pUC19 exhibits a high copy number due to the absence of the rop gene and a specific mutation in the ori region. The recognition sites for various restriction enzymes, such as HindIII, SphI, PstI, SalI, XbaI, BamHI, SmaI, KpnI, SacI, and EcoRI, have been incorporated into pUC19 from the vector M13mp19.
- The primary function of pUC19 is to introduce the plasmid into bacterial cells through a process called transformation, enabling its replication and expression. The MCS within the vector allows for the insertion of a desired foreign DNA fragment. Cells that successfully take up the plasmid can be distinguished from those that do not by their ability to grow on ampicillin-containing media. Only cells carrying the ampicillin resistance gene will survive. Furthermore, recombinant cells containing the gene of interest can be differentiated from non-recombinant cells by the color of the colonies they form on agar media supplemented with IPTG and X-gal. Recombinants appear white, while non-recombinants exhibit a blue coloration.
- The mechanism underlying pUC19’s functionality involves the lacZ fragment and its complementation with a defective form of β-galactosidase enzyme encoded by the host chromosome. The addition of IPTG to the growth medium induces the synthesis of both enzyme fragments. When grown on media supplemented with X-gal, these fragments together hydrolyze the compound, resulting in blue-colored colonies.
- However, insertion of foreign DNA into the MCS disrupts the lacZ gene, leading to insertional inactivation and the loss of intra-allelic complementation. Consequently, bacteria carrying recombinant plasmids in the MCS cannot hydrolyze X-gal, resulting in the formation of white colonies. This distinct phenotype allows for the easy identification of recombinant cells on culture media compared to non-recombinant cells, which retain the ability to hydrolyze X-gal and produce blue colonies.
- To facilitate the selection and differentiation of recombinant cells, culture media containing ampicillin, IPTG, and X-gal are typically used when working with pUC19.
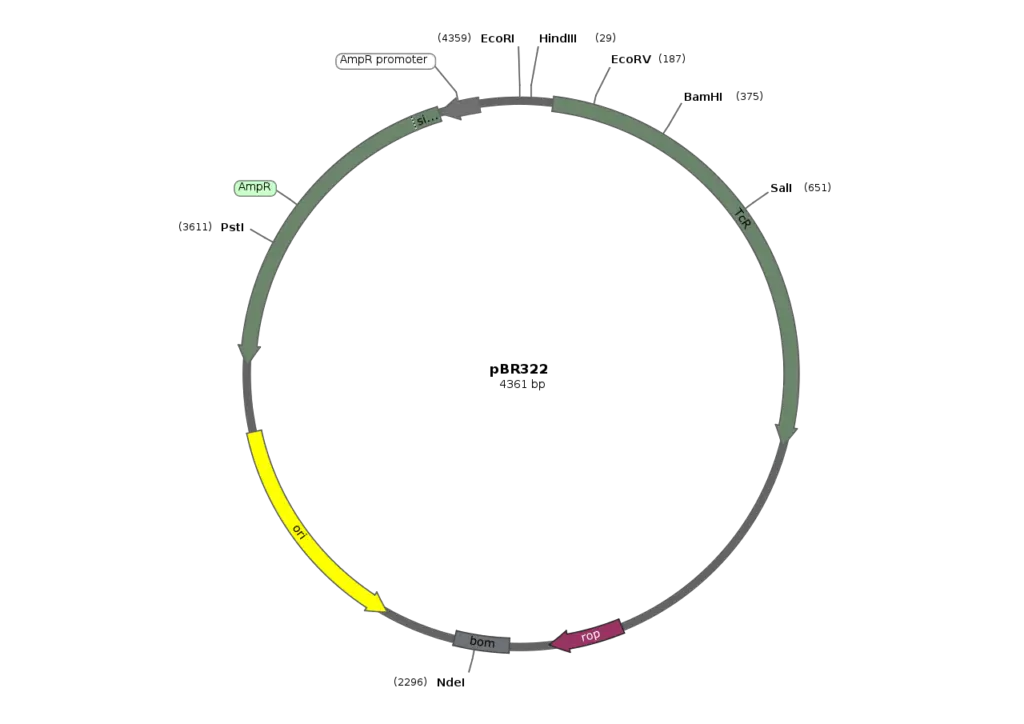
pBR322
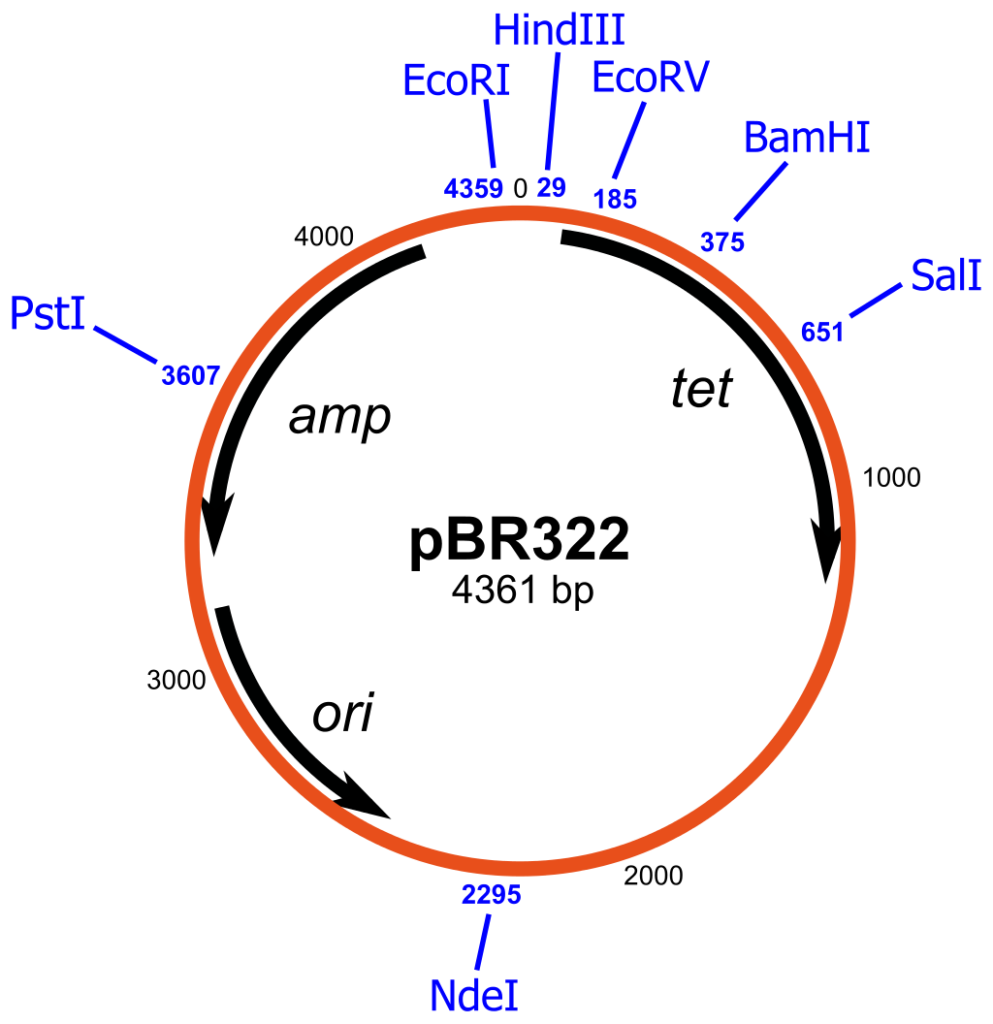
- pBR322 is a widely used plasmid cloning vector primarily employed in prokaryotes, with Escherichia coli (E. coli) being the most commonly used host organism. As a vector, pBR322 serves as a carrier for the insertion and replication of foreign DNA in bacterial cells. It was designed to address the limitations of previous vectors, such as pBR312 and pBR313, which contained extraneous DNA sequences and restriction enzyme cleavage sites that hampered their effectiveness.
- The structure of pBR322 was carefully engineered to maximize the number of restriction enzyme cleavage sites while minimizing its size. The vector consists of an origin of replication derived from the ColE1-like plasmid, pMB1, along with an ampicillin resistance gene (ApR) from the transposon Tn3 and a tetracycline resistance gene (TcR) from pSC101. These antibiotic resistance genes allow for the selection and maintenance of plasmids in bacteria that have been transformed with pBR322.
- One of the notable features of pBR322 is its inclusion of twenty-one unique restriction enzyme cleavage sites, eleven of which are found within the TcR and ApR genes. The vector also incorporates a unique EcoRI cleavage site within the plasmid, enhancing its efficiency as a vector. This extensive array of restriction enzyme cleavage sites enables researchers to easily manipulate the vector for cloning purposes.
- Although pBR322 has been a reliable and versatile cloning vector for several decades, it does have some limitations. In continuous culture without selective pressure, the vector can be lost, which can be problematic for large-scale fermentation of recombinant bacteria. Additionally, while the vector was initially designed for general cloning purposes in E. coli and similar prokaryotes, derivatives of pBR322 have been developed over time to cater to specific organisms or functions.
- pBR322, named after Francisco Bolivar Zapata and Raymond L. Rodriguez, was created in 1977 by Herbert Boyer at the University of California, San Francisco. It played a pivotal role in the early days of genetic engineering and served as a foundational tool for molecular biology research.
- In summary, pBR322 is a widely used plasmid cloning vector in prokaryotes, particularly E. coli. Its carefully designed structure, incorporating multiple restriction enzyme cleavage sites and antibiotic resistance genes, allows for efficient cloning and selection of transformed bacteria. While it has been a reliable vector, it also has certain limitations that researchers need to consider for specific applications.
λ phage
- λ-phage is a bacteriophage, specifically designed to infect the bacterial species Escherichia coli (E. coli). As a vector, it offers superior efficiency compared to other plasmid vectors due to its highly effective ability to enter bacterial cells and incorporate recombinant DNA into the host genome.
- This bacteriophage is composed of double-stranded DNA, containing essential elements for replication and various DNA sequences that encode regulatory and replicative proteins. The presence of an ori sequence is crucial for initiating DNA replication within the host cell.
- The replication process of λ-phage involves a combination of theta and rolling circle mechanisms. These mechanisms work together to generate a linear double-stranded DNA molecule. Following infection, the cos sequence present in the phage genome facilitates the circularization of the DNA, allowing it to persist within the host cell.
- A distinctive feature of λ-phage vector is the non-essential DNA sequences located between the two arms of the vector. During the cloning process, these non-essential sequences are replaced with the desired recombinant DNA. This enables the vector to carry and deliver the recombinant DNA into the host cell, where it can be incorporated into the bacterial genome.
- In summary, λ-phage serves as an efficient vector for genetic manipulation in E. coli. Its ability to efficiently enter bacterial cells and incorporate recombinant DNA, along with its replication mechanisms and circularization process, makes it a valuable tool for molecular biology research and genetic engineering applications.
Features of Vectors
Characteristics of vectors play a crucial role in molecular biology and genetic engineering processes. These features ensure the efficient replication, expression, and manipulation of genetic material. Here are some important characteristics of vectors:
- Origin of replication: Vectors should possess an origin of replication, allowing them to replicate autonomously within host cells. This feature ensures that the vector can be propagated and maintained in the host cell.
- Promoter: Vectors used for gene expression require a promoter sequence. Promoters drive the transcription of the transgene and other genes within the vector. This enables the production of the desired protein or RNA molecule.
- Cloning site: Vectors contain a cloning site, also known as a multiple cloning site or polylinker. This region contains multiple unique restriction enzyme recognition sites, allowing for the insertion of foreign DNA into the vector through ligation. The cloning site is essential for incorporating the desired DNA fragment into the vector.
- Genetic markers: Vectors often carry genetic markers that facilitate the identification and selection of host cells containing the vector. Common markers include antibiotic resistance genes, which allow for the survival and growth of cells that have taken up the vector in the presence of specific antibiotics.
- Epitope tags: Some vectors may include specific epitope tags that can be fused to the expressed protein. These tags enable the identification and purification of the protein using antibodies against the epitope. Examples of epitope tags include polyhistidine-tag, glutathione-S-transferase (GST) tag, and maltose binding protein (MBP) tag.
- Reporter genes: Vectors may incorporate reporter genes that produce easily detectable proteins or enzymes, indicating the successful insertion of the desired DNA fragment. Common reporter genes include lacZ-α, which generates a fragment of β-galactosidase that can be detected using a colorimetric assay, and genes encoding green fluorescent protein (GFP) or luciferase, which emit light for visualization.
- Targeting sequences: Expression vectors may include targeting sequences that direct the expressed protein to specific organelles or cellular locations. These sequences ensure proper localization and function of the protein within the cell.
- Protein purification tags: Some vectors contain tags or fusion partners that facilitate the purification of the expressed protein. These tags, such as polyhistidine-tag or GST tag, can be used for affinity chromatography purification, simplifying the downstream purification process.
- Integration capability: Certain vectors, such as viral vectors, have the ability to integrate the transgene into the host genome. This feature allows for stable and long-term expression of the inserted DNA.
- Size and isolation: Vectors should have an optimal size for efficient delivery and incorporation into host cells. Additionally, vectors should be easy to isolate and purify for subsequent experiments.
By considering these characteristic features, researchers can select and design vectors that meet their specific experimental needs. These features ensure successful cloning, gene expression, and manipulation of genetic material, facilitating advancements in molecular biology and genetic engineering.
Applications of Vectors
Vectors play a crucial role in various applications within molecular biology and genetic engineering, thanks to their simplicity, cost-effectiveness, and efficiency. These applications leverage the versatility of vectors to facilitate a range of processes and studies. Here are some significant applications of vectors in the field:
- Cloning: Vectors, particularly cloning vectors, are extensively used to transfer foreign DNA into host cells. This process allows for the replication and expression of the inserted DNA, enabling the production of specific proteins or the study of gene function.
- Genetic Engineering: Vectors are instrumental in engineering organisms for specific functions. For instance, they can be used to modify E. coli bacteria to produce insulin, a critical hormone used in diabetes treatment.
- Gene Isolation and Sequencing: Vectors aid in isolating specific gene sequences within a genome, enabling their subsequent analysis and determination of nucleotide sequences through DNA sequencing techniques.
- Study of Control and Regulatory Sequences: Vectors help researchers study and analyze control sequences and regulatory elements within genomes. These sequences govern gene expression and play crucial roles in cellular processes.
- Protein Structure and Function Studies: Cloning vectors are utilized to investigate the structure, function, and production of proteins in various organisms. This research provides valuable insights into protein properties, interactions, and potential applications.
- Phage Therapy: Bacteriophage vectors are employed in phage therapy, a promising treatment approach for bacterial infections in humans and animals. Phages specifically target and infect bacteria, offering a potential alternative to conventional antibiotics.
- Mutation Identification and Disease Diagnosis: Vectors can be utilized to identify mutations within DNA sequences, aiding in the diagnosis of gene defects associated with specific diseases. This application plays a crucial role in genetic screening and personalized medicine.
- Clinical Microbiology: Recombinant DNA technology, facilitated by vectors, has been instrumental in clinical microbiology. It has enabled the development of recombinant antigens, vaccines, and diagnostic probes for screening diseases such as HIV, HCV (Hepatitis C virus), and CMV (Cytomegalovirus).
Advantages of Vectors
Vectors offer several advantages in various fields of molecular biology and genetic engineering. Here are some key advantages of using vectors:
- Efficient DNA Transfer: Vectors serve as vehicles for the transfer of foreign DNA into host cells. They facilitate the efficient introduction and incorporation of the desired DNA sequences into the host genome, allowing for the expression and study of the inserted genes.
- Replication and Amplification: Vectors often contain origins of replication that enable them to replicate within host cells. This feature allows for the production of multiple copies of the vector and the inserted DNA, increasing the yield of the desired DNA sequences.
- Selectable Markers: Vectors frequently include selectable markers, such as antibiotic resistance genes, which enable the identification and selection of host cells that have successfully taken up the vector. This ensures that only cells containing the desired DNA are selected and cultured, simplifying the isolation and purification of the target DNA.
- Versatility: Vectors come in various types, each designed for specific applications. This versatility allows researchers to choose the most suitable vector for their specific experimental needs, whether it’s for cloning, protein expression, gene editing, or other purposes.
- Molecular Tags and Fusion Proteins: Vectors can be engineered to include molecular tags or fusion protein tags. These tags facilitate the detection, purification, and localization of the expressed proteins, aiding in their study and characterization.
- Modular Design: Many vectors have modular structures with multiple cloning sites, also known as polylinkers or multiple cloning regions. These regions contain multiple restriction enzyme recognition sites, allowing for easy insertion of the desired DNA sequences. This flexibility and modularity simplify the cloning process and enable the rapid generation of recombinant DNA constructs.
- Genetic Manipulation: Vectors provide a means for genetic manipulation, such as gene knockout, gene silencing, or gene overexpression. They enable researchers to introduce specific genetic modifications into host cells or organisms, allowing the study of gene function and the investigation of molecular mechanisms.
- Compatibility with Various Hosts: Vectors can be designed to be compatible with different host organisms, including bacteria, yeast, plants, and mammalian cells. This adaptability enables researchers to work with a wide range of organisms and systems, expanding the scope of their experiments and applications.
- Standardization and Well-Characterized Vectors: Over time, certain vectors have become widely adopted and well-characterized within the scientific community. This standardization facilitates reproducibility and enables researchers to build upon existing knowledge and methodologies.
Limitations of Vectors
While vectors have revolutionized molecular biology and genetic engineering, they do have certain limitations that researchers need to consider. Here are some key limitations associated with the use of vectors:
- Stability: Vectors may exhibit instability within host cells due to various factors, such as changes in metabolic energy, pH, and temperature. Different host genotypes can also impact vector stability. These factors can lead to the loss or rearrangement of the inserted DNA sequences, affecting the reliability and reproducibility of experiments.
- Gene Overexpression: The use of vectors for gene expression can sometimes result in the overexpression of the target gene in the host cell. This excessive expression can have adverse effects, such as cellular toxicity, altered cellular physiology, or misregulation of endogenous genes. Careful regulation and control of gene expression levels are necessary to avoid such issues.
- Complexity and Compatibility: For certain applications, a single type of vector may not be sufficient to achieve the desired outcome. Researchers may need to use multiple vectors, each with different characteristics or functionalities. This can introduce complexities and challenges during experimental design, vector construction, and analysis.
- Time and Cost: Developing more efficient vectors or optimizing existing ones requires significant time, effort, and resources. Research in vector design and improvement involves extensive experimentation, testing, and validation. Additionally, the production and purification of vectors can be costly, particularly for large-scale applications.
- Host-Specific Limitations: Some vectors are designed for specific host organisms, such as bacteria, yeast, plants, or mammalian cells. Each host system may have its own limitations, including differences in gene expression machinery, post-translational modifications, or immune responses. Researchers must carefully select the appropriate vector-host system to ensure compatibility and maximize the success of their experiments.
- Insert Size Limitations: Vectors have a limited capacity to accommodate large DNA inserts. The size of the insert that can be effectively cloned and maintained within a vector can vary depending on the vector type and the host organism. Large or complex DNA sequences may exceed the capacity of certain vectors, necessitating the use of alternative cloning strategies.
- Cell-Type Specificity: Some vectors may exhibit preferential efficiency or compatibility with specific cell types or tissues. This can restrict their applicability in certain experimental systems or limit the generalizability of findings across different cell types or organisms.

FAQ
What is a vector in molecular biology?
A vector in molecular biology refers to a DNA molecule used to carry and transfer foreign DNA sequences into host cells. It acts as a vehicle for the replication and expression of the inserted DNA.
What are the types of vectors commonly used in molecular biology?
Commonly used vectors include plasmids, viral vectors (e.g., retroviruses, adenoviruses), and artificial chromosomes (e.g., yeast artificial chromosomes, bacterial artificial chromosomes).
What is the purpose of using vectors in molecular biology?
Vectors are used to introduce foreign DNA into host cells for various purposes, such as gene cloning, gene expression, protein production, and genetic engineering.
How are vectors constructed?
Vectors are constructed by incorporating specific DNA elements, such as an origin of replication, promoter, cloning sites, and selectable markers, into a backbone DNA molecule. These elements facilitate replication, gene expression, and selection of transformed cells.
What is the role of selectable markers in vectors?
Selectable markers, often antibiotic resistance genes, allow for the identification and selection of host cells that have taken up the vector. Cells without the vector or those with failed transformations are eliminated through antibiotic selection.
How are vectors introduced into host cells?
Vectors can be introduced into host cells through various methods, including transformation (for bacteria), transfection (for mammalian cells), viral transduction (using viral vectors), or physical methods like electroporation or microinjection.
Can vectors integrate into the host cell’s genome?
Yes, some vectors, particularly viral vectors like retroviruses, have the ability to integrate into the host cell’s genome. This integration allows long-term expression of the inserted DNA in the host cell and its progeny.
What is the importance of multiple cloning sites (MCS) in vectors?
Multiple cloning sites, also known as polylinkers, are regions within a vector that contain multiple unique restriction enzyme recognition sites. These sites allow for the insertion of DNA fragments at specific locations, making it easier to clone different genes or DNA sequences into the vector.
Can vectors be used for gene therapy?
Yes, vectors, especially viral vectors, have been extensively used in gene therapy to deliver therapeutic genes into target cells to treat genetic disorders, cancer, and other diseases.
Are there any safety considerations when working with vectors?
Yes, safety precautions must be followed when working with vectors, particularly those derived from pathogenic sources or containing potentially harmful genes. Adherence to biosafety guidelines and proper containment measures is crucial to minimize the risk of accidental release or exposure.
References
- Gibson, D. G. (2011). Enzymatic assembly of overlapping DNA fragments. Methods in enzymology, 498, 349-361.
- Sambrook, J., Fritsch, E. F., & Maniatis, T. (1989). Molecular cloning: a laboratory manual (2nd ed.). Cold Spring Harbor Laboratory Press.
- Galloway, D. R. (2014). Introduction to cloning vectors. In Methods in molecular biology (Vol. 1116, pp. 1-12). Humana Press.
- Botstein, D., & Shortle, D. (1985). Strategies and applications of in vitro mutagenesis. Science, 229(4719), 1193-1201.
- Wirth, D., & Gama-Norton, L. (2016). Genome integration and gene targeting by homologous recombination. In Gene editing (pp. 37-61). Humana Press.
- Zinder, N. D., & Lederberg, J. (1952). Genetic exchange in Salmonella. Journal of bacteriology, 64(5), 679-699.
- Brantl, S. (2014). Plasmid replication control: replication initiator complexes and beyond. Nucleic acids research, 42(7), 3884-3893.
- Ellis, H. M., Yu, D., DiTizio, T., & Court, D. L. (2001). High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proceedings of the National Academy of Sciences, 98(12), 6742-6746.
- Muzyczka, N. (1992). Use of adeno-associated virus as a general transduction vector for mammalian cells. Current topics in microbiology and immunology, 158, 97-129.
- Weber, W., & Fussenegger, M. (2006). Molecular diversity—the toolbox for synthetic gene switches and networks. Current opinion in chemical biology, 10(6), 638-646.
